INTRODUCING-EUROFINS-BEACON-DISCOVERY-The-GPCR-Experts.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Strategies to Increase ß-Cell Mass Expansion
This electronic thesis or dissertation has been downloaded from the King’s Research Portal at https://kclpure.kcl.ac.uk/portal/ Strategies to increase -cell mass expansion Drynda, Robert Lech Awarding institution: King's College London The copyright of this thesis rests with the author and no quotation from it or information derived from it may be published without proper acknowledgement. END USER LICENCE AGREEMENT Unless another licence is stated on the immediately following page this work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International licence. https://creativecommons.org/licenses/by-nc-nd/4.0/ You are free to copy, distribute and transmit the work Under the following conditions: Attribution: You must attribute the work in the manner specified by the author (but not in any way that suggests that they endorse you or your use of the work). Non Commercial: You may not use this work for commercial purposes. No Derivative Works - You may not alter, transform, or build upon this work. Any of these conditions can be waived if you receive permission from the author. Your fair dealings and other rights are in no way affected by the above. Take down policy If you believe that this document breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 Strategies to increase β-cell mass expansion A thesis submitted by Robert Drynda For the degree of Doctor of Philosophy from King’s College London Diabetes Research Group Division of Diabetes & Nutritional Sciences Faculty of Life Sciences & Medicine King’s College London 2017 Table of contents Table of contents ................................................................................................. -

Single-Cell Rnaseq Reveals Seven Classes of Colonic Sensory Neuron
Gut Online First, published on February 26, 2018 as 10.1136/gutjnl-2017-315631 Neurogastroenterology ORIGINAL ARTICLE Gut: first published as 10.1136/gutjnl-2017-315631 on 26 February 2018. Downloaded from Single-cell RNAseq reveals seven classes of colonic sensory neuron James R F Hockley,1,2 Toni S Taylor,1 Gerard Callejo,1 Anna L Wilbrey,2 Alex Gutteridge,2 Karsten Bach,1 Wendy J Winchester,2 David C Bulmer,1 Gordon McMurray,2 Ewan St John Smith1 ► Additional material is ABSTRact pathways to the central nervous system (CNS).1 In published online only. To view Objective Integration of nutritional, microbial and the colorectum, sensory innervation is organised please visit the journal online (http:// dx. doi. org/ 10. 1136/ inflammatory events along the gut-brain axis can alter into two main pathways: thoracolumbar (TL) spinal gutjnl- 2017- 315631). bowel physiology and organism behaviour. Colonic afferents projecting via the lumbar splanchnic sensory neurons activate reflex pathways and give nerve (LSN) and lumbosacral (LS) spinal afferents 1Department of Pharmacology, University of Cambridge, rise to conscious sensation, but the diversity and projecting via the pelvic nerve (PN) that are respon- Cambridge, UK division of function within these neurons is poorly sible for transducing conscious sensations of full- 2Neuroscience and Pain understood. The identification of signalling pathways ness, discomfort, urgency and pain, in addition to Research Unit, Pfizer, contributing to visceral sensation is constrained by a reflex actions.2 Cambridge, UK paucity of molecular markers. Here we address this by Visceral sensory afferents act to maintain many comprehensive transcriptomic profiling and unsupervised aspects of GI physiology, such as continence and Correspondence to James R F Hockley, Department clustering of individual mouse colonic sensory neurons. -

F2RL2 Antibody Cat
F2RL2 Antibody Cat. No.: 56-323 F2RL2 Antibody F2RL2 Antibody immunohistochemistry analysis in formalin fixed and paraffin embedded human heart tissue followed by peroxidase conjugation of the secondary antibody and DAB staining. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human This F2RL2 antibody is generated from rabbits immunized with a KLH conjugated IMMUNOGEN: synthetic peptide between 21-50 amino acids from the N-terminal region of human F2RL2. TESTED APPLICATIONS: IHC-P, WB For WB starting dilution is: 1:1000 APPLICATIONS: For IHC-P starting dilution is: 1:10~50 PREDICTED MOLECULAR 43 kDa WEIGHT: September 25, 2021 1 https://www.prosci-inc.com/f2rl2-antibody-56-323.html Properties This antibody is purified through a protein A column, followed by peptide affinity PURIFICATION: purification. CLONALITY: Polyclonal ISOTYPE: Rabbit Ig CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: Supplied in PBS with 0.09% (W/V) sodium azide. CONCENTRATION: batch dependent Store at 4˚C for three months and -20˚C, stable for up to one year. As with all antibodies STORAGE CONDITIONS: care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: F2RL2 Proteinase-activated receptor 3, PAR-3, Coagulation factor II receptor-like 2, Thrombin ALTERNATE NAMES: receptor-like 2, F2RL2, PAR3 ACCESSION NO.: O00254 GENE ID: 2151 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References Coagulation factor II (thrombin) receptor-like 2 (F2RL2) is a member of the large family of 7-transmembrane-region receptors that couple to guanosine-nucleotide-binding proteins. -

The Orphan Receptor GPR17 Is Unresponsive to Uracil Nucleotides and Cysteinyl Leukotrienes S
Supplemental material to this article can be found at: http://molpharm.aspetjournals.org/content/suppl/2017/03/02/mol.116.107904.DC1 1521-0111/91/5/518–532$25.00 https://doi.org/10.1124/mol.116.107904 MOLECULAR PHARMACOLOGY Mol Pharmacol 91:518–532, May 2017 Copyright ª 2017 by The American Society for Pharmacology and Experimental Therapeutics The Orphan Receptor GPR17 Is Unresponsive to Uracil Nucleotides and Cysteinyl Leukotrienes s Katharina Simon, Nicole Merten, Ralf Schröder, Stephanie Hennen, Philip Preis, Nina-Katharina Schmitt, Lucas Peters, Ramona Schrage,1 Celine Vermeiren, Michel Gillard, Klaus Mohr, Jesus Gomeza, and Evi Kostenis Molecular, Cellular and Pharmacobiology Section, Institute of Pharmaceutical Biology (K.S., N.M., Ral.S., S.H., P.P., N.-K.S, L.P., J.G., E.K.), Research Training Group 1873 (K.S., E.K.), Pharmacology and Toxicology Section, Institute of Pharmacy (Ram.S., K.M.), University of Bonn, Bonn, Germany; UCB Pharma, CNS Research, Braine l’Alleud, Belgium (C.V., M.G.). Downloaded from Received December 16, 2016; accepted March 1, 2017 ABSTRACT Pairing orphan G protein–coupled receptors (GPCRs) with their using eight distinct functional assay platforms based on label- cognate endogenous ligands is expected to have a major im- free pathway-unbiased biosensor technologies, as well as molpharm.aspetjournals.org pact on our understanding of GPCR biology. It follows that the canonical second-messenger or biochemical assays. Appraisal reproducibility of orphan receptor ligand pairs should be of of GPR17 activity can be accomplished with neither the coapplica- fundamental importance to guide meaningful investigations into tion of both ligand classes nor the exogenous transfection of partner the pharmacology and function of individual receptors. -

Genome-Wide Prediction of Small Molecule Binding to Remote
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.04.236729; this version posted August 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Genome-wide Prediction of Small Molecule Binding 2 to Remote Orphan Proteins Using Distilled Sequence 3 Alignment Embedding 1 2 3 4 4 Tian Cai , Hansaim Lim , Kyra Alyssa Abbu , Yue Qiu , 5,6 1,2,3,4,7,* 5 Ruth Nussinov , and Lei Xie 1 6 Ph.D. Program in Computer Science, The Graduate Center, The City University of New York, New York, 10016, USA 2 7 Ph.D. Program in Biochemistry, The Graduate Center, The City University of New York, New York, 10016, USA 3 8 Department of Computer Science, Hunter College, The City University of New York, New York, 10065, USA 4 9 Ph.D. Program in Biology, The Graduate Center, The City University of New York, New York, 10016, USA 5 10 Computational Structural Biology Section, Basic Science Program, Frederick National Laboratory for Cancer Research, 11 Frederick, MD 21702, USA 6 12 Department of Human Molecular Genetics and Biochemistry, Sackler School of Medicine, Tel Aviv University, Tel 13 Aviv, Israel 7 14 Helen and Robert Appel Alzheimer’s Disease Research Institute, Feil Family Brain & Mind Research Institute, Weill 15 Cornell Medicine, Cornell University, New York, 10021, USA * 16 [email protected] 17 July 27, 2020 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.04.236729; this version posted August 5, 2020. -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Monitoring Nociception by Analyzing Gene Expression Changes in the Central Nervous System of Mice
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2010 Monitoring nociception by analyzing gene expression changes in the central nervous system of mice Asner, I N Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-46678 Dissertation Originally published at: Asner, I N. Monitoring nociception by analyzing gene expression changes in the central nervous system of mice. 2010, University of Zurich, Vetsuisse Faculty. Monitoring Nociception by Analyzing Gene Expression Changes in the Central Nervous System of Mice Dissertation zur Erlangung der naturwissenschaftlichen Doktorwürde (Dr. sc. nat) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultät der Universität Zürich von Igor Asner von St. Cergue VD Promotionskomitee Prof. Dr. Peter Sonderegger Prof. Dr. Kurt Bürki Prof. Dr. Hanns Ulrich Zeilhofer Dr. Paolo Cinelli (Leitung der Dissertation) Zürich, 2010 Table of contents Table of content Curriculum vitae 6 Publications 9 Summary 11 Zusammenfassung 14 1. Introduction 17 1.1. Pain and nociception 17 1.1.1 Nociceptive neurons and Mechanoceptors 18 1.1.2 Activation of the nociceptive neurons at the periphery 21 1.1.2.1 Response to noxious heat 22 1.1.2.2 Response to noxious cold 23 1.1.2.3 Response to mechanical stress 24 1.1.3 Nociceptive message processing in the Spinal Cord 25 1.1.3.1 The lamina I and the ascending pathways 25 1.1.3.2 The lamina II and the descending pathways 26 1.1.4 Pain processing and integration in the brain 27 1.1.4.1 The Pain Matrix 27 1.1.4.2 Activation of the descending pathways 29 1.1.5 Inflammatory Pain 31 1.2. -
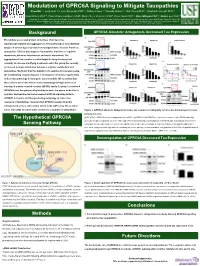
Background the Hypothetical GPRC6A Sensing Pathway
Modulation of GPRC6A Signaling to Mitigate Tauopathies Chao Ma1,2, Jerry Hunt1,3, Leslie Sandusky PhD1,3, William Fraser1,3, Ronald Alvarez1,3, Dali Zheng PhD1,4, Siddharth Kamath PhD1,2, Chad Dickey PhD1,4, Hans Bräuner-Osborne PhD5, Daniel Sejer Pedersen PhD5, Kevin Nash PhD1,2, Dave Morgan PhD1,2, Daniel Lee PhD1,3 1:Byrd Alzheimer‘s Institute, University of South Florida, Tampa, FL 33613, USA. 2:Morsani College of Medicine, Department of Molecular Pharmacology and Physiology, University of South Florida, Tampa, FL 33612, USA. 3:College of Pharmacy, Department of Pharmaceutical Sciences, University of South Florida, Tampa, FL 33612, USA. 4:Morsani College of Medicine, Department of Molecular Medicine, University of South Florida, Tampa, FL 33612, USA. 5:Department of Drug Design and Pharmacology, Faculty of Health and Medical Sciences, University of Copenhagen, Jagtvej 162, 2100 Copenhagen, Denmark Background GPRC6A Allosteric Antagonists Decreased Tau Expression Microtubule associated protein named tau, often becomes hyperphosphorylated and aggregates to form pathological neurofibrillary tangles in several age-associated neurodegenerative diseases known as tauopathies. Clinical phenotypes of tauopathies manifest as cognitive impairment, behavior disturbances and motor impairment. The aggregation of tau remains a central target for drug discovery, but currently no disease-modifying treatments exist. Our group has recently uncovered a unique interaction between L-arginine metabolism and tauopathies. We found that the depletion of L-arginine by overexpressing the metabolizing enzyme arginase 1 and arginine deiminase significantly reduced tau pathology in transgenic mouse models. We speculate that these effects associate with increased autophagy through amino acid sensing. G protein-coupled receptor (GPCR), family C, group 6, member A (GPRC6A) was deorphanized by binding to basic L-α amino acids, like L- arginine. -

G Protein-Coupled Receptors in Stem Cell Maintenance and Somatic Reprogramming to Pluripotent Or Cancer Stem Cells
BMB Reports - Manuscript Submission Manuscript Draft Manuscript Number: BMB-14-250 Title: G protein-coupled receptors in stem cell maintenance and somatic reprogramming to pluripotent or cancer stem cells Article Type: Mini Review Keywords: G protein-coupled receptors; stem cell maintenance; somatic reprogramming; cancer stem cells; pluripotent stem cell Corresponding Author: Ssang-Goo Cho Authors: Ssang-Goo Cho1,*, Hye Yeon Choi1, Subbroto Kumar Saha1, Kyeongseok Kim1, Sangsu Kim1, Gwang-Mo Yang1, BongWoo Kim1, Jin-hoi Kim1 Institution: 1Department of Animal Biotechnology, Animal Resources Research Center, and Incurable Disease Animal Model and Stem Cell Institute (IDASI), Konkuk University, 120 Neungdong-ro, Gwangjin-gu, Seoul 143-701, Republic of Korea, UNCORRECTED PROOF 1 G protein-coupled receptors in stem cell maintenance and somatic reprogramming to 2 pluripotent or cancer stem cells 3 4 Hye Yeon Choi, Subbroto Kumar Saha, Kyeongseok Kim, Sangsu Kim, Gwang-Mo 5 Yang, BongWoo Kim, Jin-hoi Kim, and Ssang-Goo Cho 6 7 Department of Animal Biotechnology, Animal Resources Research Center, and 8 Incurable Disease Animal Model and Stem Cell Institute (IDASI), Konkuk University, 9 120 Neungdong-ro, Gwangjin-gu, Seoul 143-701, Republic of Korea 10 * 11 Address correspondence to Ssang-Goo Cho, Department of Animal Biotechnology and 12 Animal Resources Research Center. Konkuk University, 120 Neungdong-ro, Gwangjin- 13 gu, Seoul 143-701, Republic of Korea. Tel: 82-2-450-4207, Fax: 82-2-450-1044, E-mail: 14 [email protected] 15 16 17 18 19 1 UNCORRECTED PROOF 20 Abstract 21 The G protein-coupled receptors (GPCRs) compose the third largest gene family in the 22 human genome, representing more than 800 distinct genes and 3–5% of the human genome. -

A Comprehensive Network and Pathway Analysis of Human Deafness Genes
Otology & Neurotology 34:961Y970 Ó 2013, Otology & Neurotology, Inc. A Comprehensive Network and Pathway Analysis of Human Deafness Genes *Georgios A. Stamatiou and †Konstantina M. Stankovic *Department of Otolaryngology, Hippokration General Hospital, University of Athens, Athens, Greece; and ÞDepartment of Otology and Laryngology, Harvard Medical School and Department of Otolaryngology, Massachusetts Eye and Ear Infirmary, Boston, Massachusetts, U.S.A. Objective: To perform comprehensive network and pathway factor beta1 (TGFB1) for Group 1, MAPK3/MAPK1 MAP kinase analyses of the genes known to cause genetic hearing loss. (ERK 1/2) and the G protein coupled receptors (GPCR) for Study Design: In silico analysis of deafness genes using inge- Group 2, and TGFB1 and hepatocyte nuclear factor 4 alpha (HNF4A) nuity pathway analysis (IPA). for Group 3. The nodal molecules included not only those known Methods: Genes relevant for hearing and deafness were iden- to be associated with deafness (GPCR), or with predisposition to tified through PubMed literature searches and the Hereditary otosclerosis (TGFB1), but also novel genes that have not been Hearing Loss Homepage. The genes were assembled into 3 groups: described in the cochlea (HNF4A) and signaling kinases (ERK 1/2). 63 genes that cause nonsyndromic deafness, 107 genes that cause Conclusion: A number of molecules that are likely to be key nonsyndromic or syndromic sensorineural deafness, and 112 genes mediators of genetic hearing loss were identified through three associated with otic capsule development and malformations. Each different network and pathway analyses. The molecules included group of genes was analyzed using IPA to discover the most new candidate genes for deafness. -

An Overview on G Protein-Coupled Receptor-Induced Signal Transduction in Acute Myeloid Leukemia
An overview on G protein-coupled receptor-induced signal transduction in Acute Myeloid Leukemia 1* 1,3 4,5,6 2* Frode Selheim , Elise Aasebø , Catalina Ribas and Anna M. Aragay 1The Proteomics Unit at the University of Bergen, Department of Biomedicine, University of Bergen, Jonas Lies vei 91, 5020 Bergen, Norway; 3 Department of Clinical Science, University of Bergen, Jonas Lies vei 87, 5021 Bergen, Norway; [email protected]. 2Departamento de Biologia Celular. Instituto de Biología Molecular de Barcelona (IBMB-CSIC), Spanish National Research Council (CSIC), Baldiri i Reixac, 15, 08028 Barcelona, Spain; [email protected]. 4Departamento de Biología Molecular and Centro de Biología Molecular “Severo Ochoa” (UAM-CSIC), 28049 Madrid, Spain; 5Instituto de Investigación Sanitaria La Princesa, 28006 Madrid, Spain; 6CIBER de Enfermedades Cardiovasculares, ISCIII (CIBERCV), 28029 Madrid, Spain, [email protected] * Corresponding authors: Frode Selheim Adr: Jonas Lies vei 91, 5020 Bergen, Norway Email: [email protected], Tel:+4755586091 Anna M. Aragay Adr: Baldiri i Reixac, 15, 08028 Barcelona. Spain. E-mail: [email protected]; Tel.: +934098671 1 Abstract Background: Acute myeloid leukemia (AML) is a genetically heterogeneous disease characterized by uncontrolled proliferation of precursor myeloid-lineage cells in the bone marrow. AML is also characterized with patients with poor long-term survival outcomes due to relapse. Many efforts have been made to understand the biological heterogeneity of AML and the challenges to develop new therapies are therefore enormous. G protein-coupled receptors (GPCRs) are a large attractive drug targeted family of transmembrane proteins, and aberrant GPCR expression and GPCR-mediated signaling have been implicated in leukemogenesis of AML.