A Minimal Mechanics Model for Mechanosensing of Substrate Rigidity Gradient in Durotaxis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
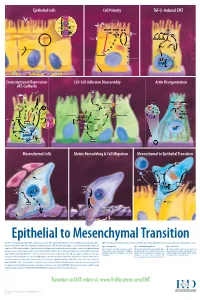
Epithelial to Mesenchymal Transition
Epithelial Cells Cell Polarity TGF-b-Induced EMT MUC-1 O-glycosylation Epithelial Cells ZO-1 Occludin Apical Membrane Tight F-Actin Microvilli Junction Claudin F-Actin p120 β-Catenin Adherens F-Actin Ezrin TGF-β dimer Junction E-Cadherin α-Catenin Plakophilin Crumbs Complex PAR Complex Desmocollin Desmoplakin Desmosome PtdIns(4,5)P2 TGF-β RII TGF-β RI CRB Cdc42Par6 Desmoglein Cytokeratin Pals1 PatJ Tight Junction Plakoglobin aPKC Par3 Domain Smad7 Extracellular PTEN JNK ERK1/2 p38 SARA Smurf1 Cortical Actin Cytoskeleton Space Par3 ZO-1 Adherens Junction PI 3-K Domain Smad-independent Signaling (–) Smad7 Translocation Smad2/3 PtdIns(3,4,5)P3 Smad4 Smad4 NEDD4 Cytokeratin Intermediate Filaments Smad2 Smad4 Smad3 LLGL Proteasome SCRIB DLG Scribble Complex Fibronectin Twist Smad2/3 Vitronectin ZEB 1/2 Microtubule Network Smad4 N-Cadherin Snail Basolateral Membrane CoA, Collagen I Slug CoR MMPs DNA-binding (+) Claudin Desmoplakin Transcription Factor Occludin Cytokeratins E-Cadherin Plakoglobin Integrins β α Nidogen-1/Entactin Perlecan Laminin Collagen IV Transcriptional Repression Cell-Cell Adhesion Disassembly Actin Reorganization of E-Cadherin TGF-β dimer EGF TGF-β RII TGF-β RI IGF FGF Receptor TNF-α Tyrosine Kinase Par6 TNF RI Apical Focal Adhesion Constriction Actin Depolymerization F-Actin Smurf1 Occludin Wnt Frizzled Myosin II Ras RhoA α-Actinin Myosin II ROCK AxinCK1 Dishevelled GSK-3 PI 3-K Src Zyxin MLC Phosphatase APC Proteasome FAK Vinculin RhoA ILK Talin (Inactive) Hakai Talin FAK F-Actin E-Cadherin LIMK Akt Paxillin FAK Stress -

A Computational Study of Stress Fiber-Focal Adhesion Dynamics
A computational study of stress fiber-focal adhesion dynamics governing cell contractility M. Maraldi1, C. Valero2, K. Garikipati1;3∗ 1Department of Mechanical Engineering, University of Michigan, Ann Arbor, Michigan 2M2BE, Aragon´ Institute of Engineering Research (I3A), University of Zaragoza, Zaragoza, Spain 3Department of Mathematics, University of Michigan, Ann Arbor, Michigan Abstract We apply a recently developed model of cytoskeletal force generation to study a cell’s intrinsic contractility, as well as its response to external loading. The model is based on a non-equilibrium thermodynamic treatment of the mechano-chemistry governing force in the stress fiber-focal adhesion system. Our computational study suggests that the mechan- ical coupling between the stress fibers and focal adhesions leads to a complex, dynamic, mechano-chemical response. We collect the results in response maps whose regimes are distinguished by the initial geometry of the stress fiber-focal adhesion system, and by the external load on the cell. The results from our model connect qualitatively with recent stud- ies on the force response of smooth muscle cells on arrays of polymeric microposts (Mann et al., Lab. on a Chip, 12, 731-740, 2012). INTRODUCTION In contractile cells, such as smooth muscle cells and fibroblasts, the generation of traction force is the result of two different actions: myosin-powered cytoskeletal contractility and external mechanical stimuli (applied stretch or force). The cooperation between these two aspects de- termines the level of the force within the cell and influences the development of cytoskeletal components via the (un)binding of proteins. Important cytoskeletal components that mediate this interplay of mechanics and chemistry are stress fibers and focal adhesions. -

Endothelial Cadherin Endothelium Is Regulated by Vascular Progenitor
Migration of Human Hematopoietic Progenitor Cells Across Bone Marrow Endothelium Is Regulated by Vascular Endothelial Cadherin This information is current as of October 1, 2021. Jaap D. van Buul, Carlijn Voermans, Veronique van den Berg, Eloise C. Anthony, Frederik P. J. Mul, Sandra van Wetering, C. Ellen van der Schoot and Peter L. Hordijk J Immunol 2002; 168:588-596; ; doi: 10.4049/jimmunol.168.2.588 http://www.jimmunol.org/content/168/2/588 Downloaded from References This article cites 54 articles, 33 of which you can access for free at: http://www.jimmunol.org/content/168/2/588.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on October 1, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2002 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Migration of Human Hematopoietic Progenitor Cells Across Bone Marrow Endothelium Is Regulated by Vascular Endothelial Cadherin1 Jaap D. van Buul,* Carlijn Voermans,* Veronique van den Berg,* Eloise C. -

Ventral Stress Fibers Induce Plasma Membrane Deformation in Human Fibroblasts
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.01.433420; this version posted March 2, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Ventral Stress Fibers Induce Plasma Membrane Deformation in Human Fibroblasts Samuel J. Ghilardi1,2, Mark S. Aronson1,2, and Allyson E. Sgro1,2 1Department of Biomedical Engineering 2Biological Design Center, Boston University, Boston, MA 02215 USA Abstract actions, actin stress fibers deform the membrane at smaller Interactions between the actin cytoskeleton and the plasma structures such as focal adhesions (27, 28) and filopodia, membrane are essential for many eukaryotic cellular processes. as well as in larger projections like lamellipodia (29–31). During these processes, actin fibers deform the cell membrane outward by applying forces parallel to the fiber’s major axis (as To deform the plasma membrane, actin stress fibers apply in migration) or they deform the membrane inward by applying force along their principle axis, either through actin polymer- forces perpendicular to the fiber’s major axis (as during cytoki- ization (32–34) or via myosin II contraction (35–38). As a nesis). Here we describe a novel actin-membrane interaction result, during most contractile events, actin stress fibers gen- in human dermal myofibroblasts. When labeled with a cytoso- erally deform the membrane parallel to the major axis of the lic fluorophore, the myofibroblasts developed prominent fluores- fiber. A notable exception to this is during specialized mem- cent structures on the ventral side of the cell. -

Cell Mechanics and Mechanobiology
CELL MECHANICS AND MECHANOBIOLOGY Hans Van Oosterwyck Biomechanics section, Department of Mechanical Engineering, KU Leuven, Belgium Liesbet Geris Biomechanics Research Unit, U.Liège, Belgium José Manuel García Aznar Multiscale in Mechanical and Biological Engineering (M2BE), Aragón Institute of Engineering Research (I3A), Universidad de Zaragoza, Spain Key Words: mechanobiology, cell mechanics, mechanotransduction, cytoskeleton, computational modelling. Contents 1. Introduction 2. Cell mechanics 2.1. Overview 2.2. Experimental techniques to measure cell mechanical properties 2.3. Computational modelling of cell mechanical properties 3. Cell mechanobiology 3.1. Overview 3.2. Mechanotransduction 3.3. Computational modelling of cell mechanobiology 4. Conclusions Acknowledgements Glossary Bibliography Biographical Sketches Summary Mechanical signals are important regulators of cell behaviour. Key to understanding their role is the fact that cells are able to sense and respond to mechanical signals. In order to unravel the interplay between mechanics and biology one needs to embrace experimental and computational methods, stemming from engineering as well as biological disciplines, and integrate them into an interdisciplinary research field called mechanobiology. In this chapter we will first describe the structural and mechanical properties of a cell and its components, as these properties will have important consequences for the way mechanical signals are converted into a biochemical response. Experimental techniques to measure and computational models to capture these properties will be highlighted. Once we have addressed some key aspects of cell mechanics, we will continue by describing some key mechanisms of how mechanical signals can modulate cell behaviour. Again, insights from experimental as well as computational studies will be reviewed. Given the broadness of the field, we will either focus on generic mechanisms, or limit ourselves to a few examples and case studies. -

A Biomechanical Perspective on Stress Fiber Structure and Function
UC Berkeley UC Berkeley Previously Published Works Title A biomechanical perspective on stress fiber structure and function. Permalink https://escholarship.org/uc/item/5w19102z Journal Biochimica et biophysica acta, 1853(11 Pt B) ISSN 0006-3002 Authors Kassianidou, Elena Kumar, Sanjay Publication Date 2015-11-01 DOI 10.1016/j.bbamcr.2015.04.006 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Biochimica et Biophysica Acta 1853 (2015) 3065–3074 Contents lists available at ScienceDirect Biochimica et Biophysica Acta journal homepage: www.elsevier.com/locate/bbamcr Review A biomechanical perspective on stress fiber structure and function☆ Elena Kassianidou, Sanjay Kumar ⁎ Department of Bioengineering, University of California, Berkeley, United States article info abstract Article history: Stress fibers are actomyosin-based bundles whose structural and contractile properties underlie numerous cellu- Received 6 January 2015 lar processes including adhesion, motility and mechanosensing. Recent advances in high-resolution live-cell im- Received in revised form 5 April 2015 aging and single-cell force measurement have dramatically sharpened our understanding of the assembly, Accepted 8 April 2015 connectivity, and evolution of various specialized stress fiber subpopulations. This in turn has motivated interest Available online 17 April 2015 in understanding how individual stress fibers generate tension and support cellular structure and force genera- fi Keywords: tion. In this review, we discuss approaches for measuring the mechanical properties of single stress bers. We Stress fiber begin by discussing studies conducted in cell-free settings, including strategies based on isolation of intact stress Biomechanical property fibers and reconstitution of stress fiber-like structures from purified components. -

Citocinas Proinflamatorias: Participación En La Modulación De La Actividad Del Melanoma Experimental B16
eman ta zabal zazu Universidad Euskal Herriko del País Vasco Unibertsitatea MEDIKUNTZA ETA ODONTOLOGIA FAKULTATEA FACULTAD DE MEDICINA Y ODONTOLOGIA Dpto. de Biología Zelulen Biologia Celular e Histología eta Histologia Saila CITOCINAS PROINFLAMATORIAS: PARTICIPACIÓN EN LA MODULACIÓN DE LA ACTIVIDAD DEL MELANOMA EXPERIMENTAL B16 Por: Juan Carlos de la Cruz Conde Licenciado en Ciencias Biológicas LEIOA, 2014 Mi más sincero agradecimiento, A la Dra. Alicia García de Galdeano, directora de este trabajo por su sinceridad, apoyo, orientación, dedicación y estímulo permanente; pero sobre todo, por la confianza que depositó en mí para la realización del mismo. La aventura que iniciamos está a punto de terminar. Juntos recorrimos todo este camino y a pesar de las adversidades, ha merecido con mucho la pena aprender y trabajar colaborando contigo en este grupo. A los Profesores Mª Luz Cañavate, Juan Aréchaga, Mª Dolores Boyano, Antonia Álvarez, Francisco José Sáez, Fernando Unda, Enrique Hilario, Gorka Pérez-Yarza, Jon Arlucea, Carmen de la Hoz y Noelia Andollo, por todas las facilidades recibidas a lo largo de la realización de esta Tesis. Entre otras: la cesión de locales y el uso de equipos técnicos (citometría de flujo y microscopía de fluorescencia) e informáticos. Por su asesoramiento científico y haber puesto a mi disposición bibliografía. Así como, material de laboratorio de todo tipo, tanto fungible como diversos productos. Además, quiero hacer mención especial al Dr. Manuel García Sanz, que aunque ya no este físicamente, nos brindó su colaboración desinteresada y del que siempre obtuvimos todo tipo de facilidades y atenciones. A mis Amigos que forman parte del Departamento de Biología Celular e Histología, en especial a Loli García Vázquez, la persona que me enseñó con cariño y paciencia infinita. -

Vinculin, and A-Actinin K.-L
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 9182-9187, August 1996 Medical Sciences The differential adhesion forces of anterior cruciate and medial collateral ligament fibroblasts: Effects of tropomodulin, talin, vinculin, and a-actinin K.-L. PAUL SUNG*tt§, LI YANG*t, DARREN E. WHITTEMOREt, YAN SHI*, GANG JIN t, ADAM H. HSIEH*t, WAYNE H. AKESON*, AND L. AMY SUNGt¶ *Departments of Orthopaedics and tBioengineering, tCancer Center, and TCenter for Molecular Genetics, Institute for Biomedical Engineering, University of California at San Diego, La Jolla, CA 92093-0412 Communicated by Y C. Fung, University of California at San Diego, La Jolla, CA, May 28, 1996 (received for review April 8, 1996) ABSTRACT We have determined the effects of tropo- fashion along the grooves of the actin double helix, stiffening modulin (Tmod), talin, vinculin, and a-actinin on ligament the filament and regulating the interaction between actin and fibroblast adhesion. The anterior cruciate ligament (ACL), other actin-binding proteins (12). Tmod may, therefore, reg- which lacks a functional healing response, and the medial ulate the length and/or organization of actin filaments by collateral ligament (MCL), a functionally healing ligament, differential binding to TM. In fibroblasts, hTM5 (one of the were selected for this study. The micropipette aspiration TM isoforms) is present not only in the more stable structures technique was used to determine the forces needed to separate of stress fibers, but also in the ruffled regions where the F-actin ACL and MCL cells from a fibronectin-coated surface. De- structures are rapidly changing (14). We found previously in a livery of exogenous tropomodulin, an actin-filament capping monocytic cell line that Tmod increases cell adhesion strength protein, into MCL fibroblasts significantly increased adhe- to fibronectin (FN), whereas the antibody against Tmod sion, whereas its monoclonal antibody (mAb) significantly decreases adhesion strength (15). -

Balance of Mechanical Forces Drives Endothelial Gap Formation And
Balance of Mechanical Forces Drives Endothelial Gap Formation and May Facilitate Cancer and Immune-Cell Extravasation Jorge Escribano, Michelle B. Chen,Emad Moeendarbary,Xuan Cao, Vivek Shenoy, Jose Manuel Garcia-Aznar, Roger D. Kamm, Fabian Spill Supporting Information, Text Detailed derivation of the model The endothelial monolayer is modeled through a number of cells in two dimensions that are connected through cell-cell adhesions. Each cell contains a number of radial contractile actin stress fibers, modeled by viscoelastic springs. Then, viscoelastic springs also represent the combined cell membrane with the adjacent cortex. For simplicity, in the paper we will refer to these elements as membrane elements. Therefore,the whole cell is discretized into nodes, which represent the fundamental degrees of freedom of the resulting network of stress fibers, membrane elements and cell-cell junctions. The cell membrane is discretized into nnodes nodes with a spacing, ln between them. The example of a hexagonal geometry is shown in Fig. S1, where all radial stress fibers are connected in the center of the cell. The actual model is independent of cellular geometry. In fact, cell geometry is dynamically changing, as observed in experiments (Movie S2), and that is reflected in the model simulations. Both membrane and stress fibers are considered as viscoelastic elements and are approximated by Kelvin-Voigt structures, in line with previous models [28]. We assume that inertial forces have no significant impact on the system, as typical on the cellular scale [54], and the forces exhibit inherent randomness due to fluctuations in molecular activities in each cell and due to variability in growth factors or neighboring cells that inevitably affects cell mechanics. -

Multiscale Model Predicts Increasing Focal Adhesion Size with Decreasing
Multiscale model predicts increasing focal adhesion PNAS PLUS size with decreasing stiffness in fibrous matrices Xuan Caoa, Ehsan Bana, Brendon M. Bakerb, Yuan Linc, Jason A. Burdickd, Christopher S. Chene, and Vivek B. Shenoya,d,1 aDepartment of Materials Science and Engineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, PA 19104; bDepartment of Biomedical Engineering, University of Michigan, Ann Arbor, MI 48109; cDepartment of Mechanical Engineering, The University of Hong Kong, Hong Kong, China; dDepartment of Bioengineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, PA 19104; and eTissue Microfabrication Laboratory, Department of Biomedical Engineering, Boston University, Boston, MA 02215 SEE COMMENTARY Edited by David A. Weitz, Harvard University, Cambridge, MA, and approved April 4, 2017 (received for review December 14, 2016) We describe a multiscale model that incorporates force-dependent on stiffer elastic substrates (17). Although these studies demon- mechanical plasticity induced by interfiber cross-link breakage and strate a clear departure from the well-described relationship be- stiffness-dependent cellular contractility to predict focal adhesion (FA) tween material stiffness and spreading established with elastic growth and mechanosensing in fibrous extracellular matrices (ECMs). hydrogel surfaces, a quantitative description of how cells are able to The model predicts that FA size depends on both the stiffness of ECM physically remodel -

Evidence of a Large-Scale Mechanosensing Mechanism for Cellular Adaptation to Substrate Stiffness
Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness Léa Tricheta,1, Jimmy Le Digabela,1, Rhoda J. Hawkinsb,c, Sri Ram Krishna Vedulad, Mukund Guptad, Claire Ribraulta, Pascal Hersena,d, Raphaël Voituriezb, and Benoît Ladouxa,d,2 aLaboratoire Matière et Systèmes Complexes, Centre National de la Recherche Scientifique Unité Mixte de Recherche, 7057, Université Paris Diderot, 75205 Paris Cedex 13, France; bCentre National de la Recherche Scientifique Unité Mixte de Recherche, 7600, Université Pierre et Marie Curie, 75252 Paris Cedex 05, France; cDepartment of Physics and Astronomy, University of Sheffield, Sheffield S3 7RH, United Kingdom; and dMechanobiology Institute, National University of Singapore, Singapore 117411 Edited by T. C. Lubensky, University of Pennsylvania, Philadelphia, PA, and approved March 6, 2012 (received for review October 28, 2011) Cell migration plays a major role in many fundamental biological modeling (18) as well as indirect observations (19, 20) suggest processes, such as morphogenesis, tumor metastasis, and wound that the contractile actomyosin apparatus can act as a global healing. As they anchor and pull on their surroundings, adhering rigidity sensor (21). From a physical point of view, the deforma- cells actively probe the stiffness of their environment. Current tion of the surrounding matrix in response to cell contractility is understanding is that traction forces exerted by cells arise mainly poorly understood; plausible mechanisms of cell mechanosensi- at mechanotransduction sites, called focal adhesions, whose size tivity imply that the regulation could be either mediated by the seems to be correlated to the force exerted by cells on their under- stress exerted by cells, or by the strain in the ECM (7, 22–24). -

Actin Stress Fibers – Assembly, Dynamics and Biological Roles
Commentary 1855 Actin stress fibers – assembly, dynamics and biological roles Sari Tojkander, Gergana Gateva and Pekka Lappalainen* Institute of Biotechnology, P.O. Box 56, University of Helsinki, 00014, Finland *Author for correspondence ([email protected]) Journal of Cell Science 125, 1855–1864 ß 2012. Published by The Company of Biologists Ltd doi: 10.1242/jcs.098087 Summary Actin filaments assemble into diverse protrusive and contractile structures to provide force for a number of vital cellular processes. Stress fibers are contractile actomyosin bundles found in many cultured non-muscle cells, where they have a central role in cell adhesion and morphogenesis. Focal-adhesion-anchored stress fibers also have an important role in mechanotransduction. In animal tissues, stress fibers are especially abundant in endothelial cells, myofibroblasts and epithelial cells. Importantly, recent live-cell imaging studies have provided new information regarding the mechanisms of stress fiber assembly and how their contractility is regulated in cells. In addition, these studies might elucidate the general mechanisms by which contractile actomyosin arrays, including muscle cell myofibrils and cytokinetic contractile ring, can be generated in cells. In this Commentary, we discuss recent findings concerning the physiological roles of stress fibers and the mechanism by which these structures are generated in cells. Key words: Actin, Adhesion, Assembly, Mechanotransduction, Stress fibers Introduction muscle cells and stress fibers of non-muscle