Cell Shape and Durotaxis Explained from Cell-Extracellular Matrix Forces and Focal Adhesion Dynamics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Development of Microfluidic Devices to Study Algal Chemotaxis and Long-Term Growth Dynamics" (2016)
Louisiana State University LSU Digital Commons LSU Master's Theses Graduate School 2016 Development of Microfluidic evD ices to Study Algal Chemotaxis and Long-Term Growth Dynamics Benjamin Seth Roberts Louisiana State University and Agricultural and Mechanical College, [email protected] Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_theses Part of the Chemical Engineering Commons Recommended Citation Roberts, Benjamin Seth, "Development of Microfluidic Devices to Study Algal Chemotaxis and Long-Term Growth Dynamics" (2016). LSU Master's Theses. 4496. https://digitalcommons.lsu.edu/gradschool_theses/4496 This Thesis is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Master's Theses by an authorized graduate school editor of LSU Digital Commons. For more information, please contact [email protected]. DEVELOPMENT OF MICROFLUIDIC DEVICES TO STUDY ALGAL CHEMOTAXIS AND LONG-TERM GROWTH DYNAMICS A Thesis Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Master of Science in The Cain Department of Chemical Engineering by Benjamin S. Roberts B.S., Mississippi State University, 2014 December 2016 TABLE OF CONTENTS ABSTRACT ................................................................................................................................... iii CHAPTER 1. INTRODUCTION ....................................................................................................1 -

Morphological Study of Cell Protrusions During Redirected Migration in Human Fibroblast Cells
MORPHOLOGICAL STUDY OF CELL PROTRUSIONS DURING REDIRECTED MIGRATION IN HUMAN FIBROBLAST CELLS Congyingzi Zhang A Thesis Submitted to the Graduate College of Bowling Green State University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE August 2013 Committee: Dr. Carol Heckman, Advisor Dr. Roudabeh Jamasbi Dr. Peter Gorsevski ii ABSTRACT Carol A. Heckman, Advisor From the perspective of cell motility mechanisms, migration patterns arise from two opposing sources which can be viewed as forces. One, called intrinsic, maintains the cell persistence. The extrinsic arises from signals (repulsive or attractive) exerted by an external stimulus. The extrinsic force is stronger than the intrinsic, since it can overcome the intrinsic force and cause the cell to change direction. The current studies were designed to determine whether these forces were associated with different protrusions. I studied human fibroblast cells that collide with a haptotactic boundary between an adhesive substrate (germanium) and a non- adhesive substrate (plastic) in a chemokinesis system. The morphologies of cells migrating on the two substrates reflected the cells’ preference for the adhesive substrate. I measured the prevalence of various protrusions during the process of cells turning away from the boundary and reorienting their direction of travel. Classes that corresponded to protrusive features were identified by extracting latent factors from a number of primary, geometric variables, and included factor 4 (filopodia), factor 5 (cell mass displacement), and factor 7 (nascent neurites). The data showed that as cells moved further and further from the boundary, they had progressively lower values of factor 5. The correlation coefficient between the values is -0.4924. -

Negative Durotaxis: Cell Movement Toward Softer Environments
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.27.357178; this version posted October 27, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Negative durotaxis: cell movement toward softer environments Aleksi Isomursu1,*, Keun-Young Park2,*, Jay Hou3,*, Bo Cheng4,5,*, Ghaidan Shamsan3, Benjamin Fuller3, Jesse Kasim3, M. Mohsen Mahmoodi2, Tian Jian Lu6,7, Guy M. Genin4,5,8, Feng Xu4,5, Min Lin4,5,#, Mark Distefano2,#, Johanna Ivaska1,9,#, and David J. Odde3,# 1Turku Bioscience Centre, University of Turku and Åbo Akademi University, 20520 Turku, Finland 2Department of Chemistry, University of Minnesota, Minneapolis 55455, MN, USA 3Department of Biomedical Engineering, University of Minnesota, Minneapolis 55455, MN, USA 4The Key Laboratory of Biomedical Information Engineering of Ministry of Education, School of Life Science and Technology, Xi’an Jiaotong University, Xi’an 710049, P.R. China 5Bioinspired Engineering and Biomechanics Center (BEBC), Xi’an Jiaotong University, Xi’an 710049, P.R. China 6State Key Laboratory of Mechanics and Control of Mechanical Structures, Nanjing University of Aeronautics and Astronautics, NanjinG 210016, P.R. China 7MOE Key Laboratory of Multifunctional Materials and Structures, Xi’an JiaotonG University, Xi’an 710049, P.R. China 8NSF Science and TechnoloGy Center for EnGineerinG MechanobioloGy, WashinGton University in St. Louis, St. Louis 63130, MO, USA 9Department of Biochemistry, University of Turku, 20520 Turku, Finland *Equal contribution #Corresponding authors Email: [email protected] (D.J.O.); [email protected] (J.I.); [email protected] (M.D.); [email protected] (M.L.) Abstract: Durotaxis – the ability of cells to sense and migrate along stiffness gradients – is important for embryonic development and has been implicated in pathologies including fibrosis and cancer. -
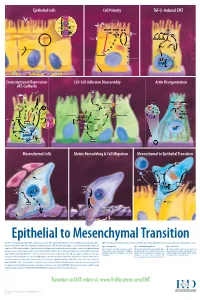
Epithelial to Mesenchymal Transition
Epithelial Cells Cell Polarity TGF-b-Induced EMT MUC-1 O-glycosylation Epithelial Cells ZO-1 Occludin Apical Membrane Tight F-Actin Microvilli Junction Claudin F-Actin p120 β-Catenin Adherens F-Actin Ezrin TGF-β dimer Junction E-Cadherin α-Catenin Plakophilin Crumbs Complex PAR Complex Desmocollin Desmoplakin Desmosome PtdIns(4,5)P2 TGF-β RII TGF-β RI CRB Cdc42Par6 Desmoglein Cytokeratin Pals1 PatJ Tight Junction Plakoglobin aPKC Par3 Domain Smad7 Extracellular PTEN JNK ERK1/2 p38 SARA Smurf1 Cortical Actin Cytoskeleton Space Par3 ZO-1 Adherens Junction PI 3-K Domain Smad-independent Signaling (–) Smad7 Translocation Smad2/3 PtdIns(3,4,5)P3 Smad4 Smad4 NEDD4 Cytokeratin Intermediate Filaments Smad2 Smad4 Smad3 LLGL Proteasome SCRIB DLG Scribble Complex Fibronectin Twist Smad2/3 Vitronectin ZEB 1/2 Microtubule Network Smad4 N-Cadherin Snail Basolateral Membrane CoA, Collagen I Slug CoR MMPs DNA-binding (+) Claudin Desmoplakin Transcription Factor Occludin Cytokeratins E-Cadherin Plakoglobin Integrins β α Nidogen-1/Entactin Perlecan Laminin Collagen IV Transcriptional Repression Cell-Cell Adhesion Disassembly Actin Reorganization of E-Cadherin TGF-β dimer EGF TGF-β RII TGF-β RI IGF FGF Receptor TNF-α Tyrosine Kinase Par6 TNF RI Apical Focal Adhesion Constriction Actin Depolymerization F-Actin Smurf1 Occludin Wnt Frizzled Myosin II Ras RhoA α-Actinin Myosin II ROCK AxinCK1 Dishevelled GSK-3 PI 3-K Src Zyxin MLC Phosphatase APC Proteasome FAK Vinculin RhoA ILK Talin (Inactive) Hakai Talin FAK F-Actin E-Cadherin LIMK Akt Paxillin FAK Stress -

Bimodal Rheotactic Behavior Reflects Flagellar Beat Asymmetry in Human Sperm Cells
Bimodal rheotactic behavior reflects flagellar beat asymmetry in human sperm cells Anton Bukatina,b,1, Igor Kukhtevichb,c,1, Norbert Stoopd,1, Jörn Dunkeld,2, and Vasily Kantslere aSt. Petersburg Academic University, St. Petersburg 194021, Russia; bInstitute for Analytical Instrumentation of the Russian Academy of Sciences, St. Petersburg 198095, Russia; cITMO University, St. Petersburg 197101, Russia; dDepartment of Mathematics, Massachusetts Institute of Technology, Cambridge, MA 02139-4307; and eDepartment of Physics, University of Warwick, Coventry CV4 7AL, United Kingdom Edited by Charles S. Peskin, New York University, New York, NY, and approved November 9, 2015 (received for review July 30, 2015) Rheotaxis, the directed response to fluid velocity gradients, has whether this effect is of mechanical (20) or hydrodynamic (21, been shown to facilitate stable upstream swimming of mamma- 22) origin. Experiments (23) show that the alga’s reorientation lian sperm cells along solid surfaces, suggesting a robust physical dynamics can lead to localization in shear flow (24, 25), with mechanism for long-distance navigation during fertilization. How- potentially profound implications in marine ecology. In contrast ever, the dynamics by which a human sperm orients itself relative to taxis in multiflagellate organisms (2, 5, 18, 26, 27), the navi- to an ambient flow is poorly understood. Here, we combine micro- gation strategies of uniflagellate cells are less well understood. fluidic experiments with mathematical modeling and 3D flagellar beat For instance, it was discovered only recently that uniflagellate reconstruction to quantify the response of individual sperm cells in marine bacteria, such as Vibrio alginolyticus and Pseudoalteromonas time-varying flow fields. Single-cell tracking reveals two kinematically haloplanktis, use a buckling instability in their lone flagellum to distinct swimming states that entail opposite turning behaviors under change their swimming direction (28). -

A Hybrid Computational Model for Collective Cell Durotaxis
Biomechanics and Modeling in Mechanobiology https://doi.org/10.1007/s10237-018-1010-2 ORIGINAL PAPER A hybrid computational model for collective cell durotaxis Jorge Escribano1 · Raimon Sunyer2,5 · María Teresa Sánchez3 · Xavier Trepat2,4,5,6 · Pere Roca-Cusachs2,4 · José Manuel García-Aznar1 Received: 13 September 2017 / Accepted: 17 February 2018 © Springer-Verlag GmbH Germany, part of Springer Nature 2018 Abstract Collective cell migration is regulated by a complex set of mechanical interactions and cellular mechanisms. Collective migration emerges from mechanisms occurring at single cell level, involving processes like contraction, polymerization and depolymerization, of cell–cell interactions and of cell–substrate adhesion. Here, we present a computational framework which simulates the dynamics of this emergent behavior conditioned by substrates with stiffness gradients. The computational model reproduces the cell’s ability to move toward the stiffer part of the substrate, process known as durotaxis. It combines the continuous formulation of truss elements and a particle-based approach to simulate the dynamics of cell–matrix adhesions and cell–cell interactions. Using this hybrid approach, researchers can quickly create a quantitative model to understand the regulatory role of different mechanical conditions on the dynamics of collective cell migration. Our model shows that durotaxis occurs due to the ability of cells to deform the substrate more in the part of lower stiffness than in the stiffer part. This effect explains why cell collective movement is more effective than single cell movement in stiffness gradient conditions. In addition, we numerically evaluate how gradient stiffness properties, cell monolayer size and force transmission between cells and extracellular matrix are crucial in regulating durotaxis. -

Engineering 3D Systems with Tunable Mechanical Properties to Mimic the Tumor Microenvironment Shalini Raj Unnikandam Veettil Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2018 Engineering 3D systems with tunable mechanical properties to mimic the tumor microenvironment Shalini Raj Unnikandam Veettil Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Chemical Engineering Commons Recommended Citation Unnikandam Veettil, Shalini Raj, "Engineering 3D systems with tunable mechanical properties to mimic the tumor microenvironment" (2018). Graduate Theses and Dissertations. 17339. https://lib.dr.iastate.edu/etd/17339 This Thesis is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Engineering 3D systems with tunable mechanical properties to mimic the tumor microenvironment by Shalini Raj Unnikandam Veettil A thesis submitted to the graduate faculty in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE Major: Chemical Engineering Program of Study Committee: Ian C Schneider, Major Professor Kaitlin Bratlie Michael Bartlett The student author, whose presentation of the scholarship herein was approved by the program of study committee, is solely responsible for the content of this thesis. The Graduate College will ensure this thesis is globally accessible and will not permit alterations after a degree is conferred. Iowa State University Ames, Iowa 2018 Copyright © Shalini Raj Unnikandam Veettil, 2018. All rights reserved. ii DEDICATION This thesis is dedicated to my family and friends who have been a great source of support. -

A Computational Study of Stress Fiber-Focal Adhesion Dynamics
A computational study of stress fiber-focal adhesion dynamics governing cell contractility M. Maraldi1, C. Valero2, K. Garikipati1;3∗ 1Department of Mechanical Engineering, University of Michigan, Ann Arbor, Michigan 2M2BE, Aragon´ Institute of Engineering Research (I3A), University of Zaragoza, Zaragoza, Spain 3Department of Mathematics, University of Michigan, Ann Arbor, Michigan Abstract We apply a recently developed model of cytoskeletal force generation to study a cell’s intrinsic contractility, as well as its response to external loading. The model is based on a non-equilibrium thermodynamic treatment of the mechano-chemistry governing force in the stress fiber-focal adhesion system. Our computational study suggests that the mechan- ical coupling between the stress fibers and focal adhesions leads to a complex, dynamic, mechano-chemical response. We collect the results in response maps whose regimes are distinguished by the initial geometry of the stress fiber-focal adhesion system, and by the external load on the cell. The results from our model connect qualitatively with recent stud- ies on the force response of smooth muscle cells on arrays of polymeric microposts (Mann et al., Lab. on a Chip, 12, 731-740, 2012). INTRODUCTION In contractile cells, such as smooth muscle cells and fibroblasts, the generation of traction force is the result of two different actions: myosin-powered cytoskeletal contractility and external mechanical stimuli (applied stretch or force). The cooperation between these two aspects de- termines the level of the force within the cell and influences the development of cytoskeletal components via the (un)binding of proteins. Important cytoskeletal components that mediate this interplay of mechanics and chemistry are stress fibers and focal adhesions. -

Endothelial Cadherin Endothelium Is Regulated by Vascular Progenitor
Migration of Human Hematopoietic Progenitor Cells Across Bone Marrow Endothelium Is Regulated by Vascular Endothelial Cadherin This information is current as of October 1, 2021. Jaap D. van Buul, Carlijn Voermans, Veronique van den Berg, Eloise C. Anthony, Frederik P. J. Mul, Sandra van Wetering, C. Ellen van der Schoot and Peter L. Hordijk J Immunol 2002; 168:588-596; ; doi: 10.4049/jimmunol.168.2.588 http://www.jimmunol.org/content/168/2/588 Downloaded from References This article cites 54 articles, 33 of which you can access for free at: http://www.jimmunol.org/content/168/2/588.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on October 1, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2002 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Migration of Human Hematopoietic Progenitor Cells Across Bone Marrow Endothelium Is Regulated by Vascular Endothelial Cadherin1 Jaap D. van Buul,* Carlijn Voermans,* Veronique van den Berg,* Eloise C. -

Ventral Stress Fibers Induce Plasma Membrane Deformation in Human Fibroblasts
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.01.433420; this version posted March 2, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Ventral Stress Fibers Induce Plasma Membrane Deformation in Human Fibroblasts Samuel J. Ghilardi1,2, Mark S. Aronson1,2, and Allyson E. Sgro1,2 1Department of Biomedical Engineering 2Biological Design Center, Boston University, Boston, MA 02215 USA Abstract actions, actin stress fibers deform the membrane at smaller Interactions between the actin cytoskeleton and the plasma structures such as focal adhesions (27, 28) and filopodia, membrane are essential for many eukaryotic cellular processes. as well as in larger projections like lamellipodia (29–31). During these processes, actin fibers deform the cell membrane outward by applying forces parallel to the fiber’s major axis (as To deform the plasma membrane, actin stress fibers apply in migration) or they deform the membrane inward by applying force along their principle axis, either through actin polymer- forces perpendicular to the fiber’s major axis (as during cytoki- ization (32–34) or via myosin II contraction (35–38). As a nesis). Here we describe a novel actin-membrane interaction result, during most contractile events, actin stress fibers gen- in human dermal myofibroblasts. When labeled with a cytoso- erally deform the membrane parallel to the major axis of the lic fluorophore, the myofibroblasts developed prominent fluores- fiber. A notable exception to this is during specialized mem- cent structures on the ventral side of the cell. -

Phenomenlogical Modeling of Durotaxis
Phenomenlogical Modeling of Durotaxis Guangyuan Yu1;2, Jingchen Feng2, Haoran Man1,Herbert Levine2;3 1 Physics and Astronomy Department, Rice University, Houston, USA 2 Center for Theoretical Biological Physics, Rice University, Houston, USA 3 Bioengineering Department, Rice University, Houston, USA (Dated: February 23, 2017) Cells exhibit qualitatively different behaviors on substrates with different rigidities. The fact that cells are more polarized on the stiffer substrate motivates us to construct a two-dimensional cell with the distribution of focal adhesions dependent on substrate rigidities. This distribution affects the forces exerted by the cell and thereby determines its motion. Our model reproduces the experimental observation that the persistence time is higher on the stiffer substrate. This stiffness dependent persistence will lead to durotaxis, the preference in moving towards stiffer substrates. This propensity is characterized by the durotaxis index first defined in experiments. We derive and validate a 2D corresponding Fokker-Planck equation associated with our model. Our approach highlights the possible role of the focal adhesion arrangement in durotaxis. Cells are capable of sensing and responding to the me- sense a stiffer substrate, they take on a more elongated chanical properties of their external environment. For shape [25, 26] as a response. Now, cells move by protru- example, cytoskeletal stiffness [1], cellular differentia- sion which occurs with the help of focal adhesions which tion [2–5] and cell morphology and motility [6–8] are all allow force transmission to the substrate. We will assume strongly influenced by ECM stiffness. In particular, it that the change in shape to being more polarized implies has been shown experimentally that cells prefer crawl- that focal adhesions (FA) are formed within a narrower ing towards the stiffer parts on substrates with spatially wedge on the cell front. -

Hipocrate 460BC–370 BC
L31 - Motility or Morbidity in Neurosurgery Motto: „I move, therefore I am” (Haruki Murakami, Japanese Writer) Alexandru Vlad Ciurea1,2 MD., PhD., MSc., Dr. h.c. Mult. Prof. Alexandru Vlad Ciurea – e-mail: [email protected] Balneo Research Journal DOI: http://dx.doi.org/10.12680/balneo.2019.276 R Vol.10, No.3, September 2019 p: 353-354 1: Department of Neurosurgery, Carol Davila University School of Medicine, Bucharest, Romania. 2: Department of Neurosurgery, Sanador Medical Center Hospital, Bucharest, Romania. Abstract Background : „The lack of motion is a basic process in the birth of diseases”. (Hipocrate 460BC–370 BC) Biological motility is the capacity of an organism to move spontaneously and voluntary at the expense of energy and represents one of the crucial traits (if not the most important trait) of all living matter. Motility is a key process present in unicellular organisms as well as multicellular organisms and is the basis of embryonic development, would healing, immune response, tumor formation, metastasis, cell migration, digestion, circulation, respiration and reasoning and various other critical processes including biological evolution itself. The constant motility of developing organisms (from trilobites to human beings) has undoubtedly shaped evolution by constantly improving the functions of the skeletal system and other internal organs such as the digestive tract, liver, kidneys etc. The evolution from prehistoric human beings to modern men went through several steps during which human motility represented once more a crucial element. Once industrial food production appeared – with the overproduction we are familiarized with, human motility became a protection factor against obesity which is one of the most problematic medical conditions of our time.