[3H]-Piclamilast and [3H]-Rolipram
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download Product Insert (PDF)
PRODUCT INFORMATION Piclamilast Item No. 29170 CAS Registry No.: 144035-83-6 Formal Name: 3-(cyclopentyloxy)-N-(3,5-dichloro- 4-pyridinyl)-4-methoxy-benzamide O Synonyms: RP 73401, RPR 73401 Cl H MF: C18H18Cl2N2O3 FW: 381.3 N O Purity: ≥98% O Supplied as: A solid N Storage: -20°C Cl Stability: ≥2 years Information represents the product specifications. Batch specific analytical results are provided on each certificate of analysis. Laboratory Procedures Piclamilast is supplied as a solid. A stock solution may be made by dissolving the piclamilast in the solvent of choice, which should be purged with an inert gas. Piclamilast is soluble in organic solvents such as ethanol and DMSO. The solubility of piclamilast in ethanol is approximately 20 mM and approximately 100 mM in DMSO. Description 1 Piclamilast is a phosphodiesterase 4 (PDE4) inhibitor (IC50 = 0.31 nM). It is selective for PDE4 over PDE1, 2,3 PDE2, PDE3, PDE5, and PDE7A in cell-free assays (IC50s = >100, 40, >100, 14, and >10 μM, respectively). Piclamilast inhibits superoxide production by guinea pig eosinophils and histamine-induced contraction 1,4 in isolated guinea pig trachea (IC50s = 24 and 2 nM, respectively). It inhibits eosinophil, neutrophil, lymphocyte, and TNF-α accumulation in bronchoalveolar lavage fluid (BALF) from ovalbumin-sensitized 5 rats (ED50s = 23.8, 14.1, 19.5, and 14.4 μmol/kg, respectively). Piclamilast also inhibits ovalbumin-induced 2 bronchoconstriction in a guinea pig model of asthma (ED50 = 0.033 mg/kg). References 1. Ashton, M.J., Cook, D.C., Fenton, G., et al. Selective type IV phosphodiesterase inhibitors as antiasthmatic agents. -

The Single Cyclic Nucleotide-Specific Phosphodiesterase of the Intestinal Parasite Giardia Lamblia Represents a Potential Drug Target
RESEARCH ARTICLE The single cyclic nucleotide-specific phosphodiesterase of the intestinal parasite Giardia lamblia represents a potential drug target Stefan Kunz1,2*, Vreni Balmer1, Geert Jan Sterk2, Michael P. Pollastri3, Rob Leurs2, Norbert MuÈ ller1, Andrew Hemphill1, Cornelia Spycher1¤ a1111111111 1 Institute of Parasitology, Vetsuisse Faculty, University of Bern, Bern, Switzerland, 2 Division of Medicinal Chemistry, Faculty of Sciences, Amsterdam Institute of Molecules, Medicines and Systems (AIMMS), Vrije a1111111111 Universiteit Amsterdam, Amsterdam, The Netherlands, 3 Department of Chemistry and Chemical Biology, a1111111111 Northeastern University, Boston, Massachusetts, United States of America a1111111111 a1111111111 ¤ Current address: Euresearch, Head Office Bern, Bern, Switzerland * [email protected] Abstract OPEN ACCESS Citation: Kunz S, Balmer V, Sterk GJ, Pollastri MP, Leurs R, MuÈller N, et al. (2017) The single cyclic Background nucleotide-specific phosphodiesterase of the Giardiasis is an intestinal infection correlated with poverty and poor drinking water quality, intestinal parasite Giardia lamblia represents a potential drug target. PLoS Negl Trop Dis 11(9): and treatment options are limited. According to the Center for Disease Control and Preven- e0005891. https://doi.org/10.1371/journal. tion, Giardia infections afflict nearly 33% of people in developing countries, and 2% of the pntd.0005891 adult population in the developed world. This study describes the single cyclic nucleotide- Editor: Aaron R. Jex, University of Melbourne, specific phosphodiesterase (PDE) of G. lamblia and assesses PDE inhibitors as a new gen- AUSTRALIA eration of anti-giardial drugs. Received: December 5, 2016 Accepted: August 21, 2017 Methods Published: September 15, 2017 An extensive search of the Giardia genome database identified a single gene coding for a class I PDE, GlPDE. -

Chapter Introduction
VU Research Portal Trypanosoma brucei phosphodiesterase B1 as a drug target for Human African Trypanosomiasis Jansen, C.J.W. 2015 document version Publisher's PDF, also known as Version of record Link to publication in VU Research Portal citation for published version (APA) Jansen, C. J. W. (2015). Trypanosoma brucei phosphodiesterase B1 as a drug target for Human African Trypanosomiasis. General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. E-mail address: [email protected] Download date: 05. Oct. 2021 An introduction into phosphodiesterases and their potential role as drug targets for neglected diseases Chapter 1 4 CHAPTER 1 1.1 Human African Trypanosomiasis Human African Trypanomiasis (HAT), also known as African sleeping sickness, is a deadly infectious disease caused by the kinetoplastid Trypanosoma -

Signal Transduction Guide
Signal Transduction Product Guide | 2007 NEW! Selective T-type Ca2+ channel blockers, NNC 55-0396 and Mibefradil ZM 447439 – Novel Aurora Kinase Inhibitor NEW! Antibodies for Cancer Research EGFR-Kinase Selective Inhibitors – BIBX 1382 and BIBU 1361 DRIVING RESEARCH FURTHER Calcium Signaling Agents ...................................2 G Protein Reagents ...........................................12 Cell Cycle and Apoptosis Reagents .....................3 Ion Channel Modulators ...................................13 Cyclic Nucleotide Related Tools ...........................7 Lipid Signaling Agents ......................................17 Cytokine Signaling Agents ..................................9 Nitric Oxide Tools .............................................19 Enzyme Inhibitors/Substrates/Activators ..............9 Protein Kinase Reagents....................................22 Glycobiology Agents .........................................12 Protein Phosphatase Reagents ..........................33 Neurochemicals | Signal Transduction Agents | Peptides | Biochemicals Signal Transduction Product Guide Calcium Signaling Agents ......................................................................................................................2 Calcium Binding Protein Modulators ...................................................................................................2 Calcium ATPase Modulators .................................................................................................................2 Calcium Sensitive Protease -

Stems for Nonproprietary Drug Names
USAN STEM LIST STEM DEFINITION EXAMPLES -abine (see -arabine, -citabine) -ac anti-inflammatory agents (acetic acid derivatives) bromfenac dexpemedolac -acetam (see -racetam) -adol or analgesics (mixed opiate receptor agonists/ tazadolene -adol- antagonists) spiradolene levonantradol -adox antibacterials (quinoline dioxide derivatives) carbadox -afenone antiarrhythmics (propafenone derivatives) alprafenone diprafenonex -afil PDE5 inhibitors tadalafil -aj- antiarrhythmics (ajmaline derivatives) lorajmine -aldrate antacid aluminum salts magaldrate -algron alpha1 - and alpha2 - adrenoreceptor agonists dabuzalgron -alol combined alpha and beta blockers labetalol medroxalol -amidis antimyloidotics tafamidis -amivir (see -vir) -ampa ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) subgroup: -ampanel antagonists becampanel -ampator modulators forampator -anib angiogenesis inhibitors pegaptanib cediranib 1 subgroup: -siranib siRNA bevasiranib -andr- androgens nandrolone -anserin serotonin 5-HT2 receptor antagonists altanserin tropanserin adatanserin -antel anthelmintics (undefined group) carbantel subgroup: -quantel 2-deoxoparaherquamide A derivatives derquantel -antrone antineoplastics; anthraquinone derivatives pixantrone -apsel P-selectin antagonists torapsel -arabine antineoplastics (arabinofuranosyl derivatives) fazarabine fludarabine aril-, -aril, -aril- antiviral (arildone derivatives) pleconaril arildone fosarilate -arit antirheumatics (lobenzarit type) lobenzarit clobuzarit -arol anticoagulants (dicumarol type) dicumarol -

Phosphodiesterase (PDE)
Phosphodiesterase (PDE) Phosphodiesterase (PDE) is any enzyme that breaks a phosphodiester bond. Usually, people speaking of phosphodiesterase are referring to cyclic nucleotide phosphodiesterases, which have great clinical significance and are described below. However, there are many other families of phosphodiesterases, including phospholipases C and D, autotaxin, sphingomyelin phosphodiesterase, DNases, RNases, and restriction endonucleases, as well as numerous less-well-characterized small-molecule phosphodiesterases. The cyclic nucleotide phosphodiesterases comprise a group of enzymes that degrade the phosphodiester bond in the second messenger molecules cAMP and cGMP. They regulate the localization, duration, and amplitude of cyclic nucleotide signaling within subcellular domains. PDEs are therefore important regulators ofsignal transduction mediated by these second messenger molecules. www.MedChemExpress.com 1 Phosphodiesterase (PDE) Inhibitors, Activators & Modulators (+)-Medioresinol Di-O-β-D-glucopyranoside (R)-(-)-Rolipram Cat. No.: HY-N8209 ((R)-Rolipram; (-)-Rolipram) Cat. No.: HY-16900A (+)-Medioresinol Di-O-β-D-glucopyranoside is a (R)-(-)-Rolipram is the R-enantiomer of Rolipram. lignan glucoside with strong inhibitory activity Rolipram is a selective inhibitor of of 3', 5'-cyclic monophosphate (cyclic AMP) phosphodiesterases PDE4 with IC50 of 3 nM, 130 nM phosphodiesterase. and 240 nM for PDE4A, PDE4B, and PDE4D, respectively. Purity: >98% Purity: 99.91% Clinical Data: No Development Reported Clinical Data: No Development Reported Size: 1 mg, 5 mg Size: 10 mM × 1 mL, 10 mg, 50 mg (R)-DNMDP (S)-(+)-Rolipram Cat. No.: HY-122751 ((+)-Rolipram; (S)-Rolipram) Cat. No.: HY-B0392 (R)-DNMDP is a potent and selective cancer cell (S)-(+)-Rolipram ((+)-Rolipram) is a cyclic cytotoxic agent. (R)-DNMDP, the R-form of DNMDP, AMP(cAMP)-specific phosphodiesterase (PDE) binds PDE3A directly. -

Inhibition of Phosphodiesterase 4 Modulates Cytokine Induction From
University of Groningen Inhibition of phosphodiesterase 4 modulates cytokine induction from toll like receptor activated, but not rhinovirus infected, primary human airway smooth muscle Van Ly, David; De Pedro, Monique; James, Peter; Morgan, Lucy; Black, Judith L.; Burgess, Janette K.; Oliver, Brian G. G. Published in: Respiratory Research DOI: 10.1186/1465-9921-14-127 IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2013 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Van Ly, D., De Pedro, M., James, P., Morgan, L., Black, J. L., Burgess, J. K., & Oliver, B. G. G. (2013). Inhibition of phosphodiesterase 4 modulates cytokine induction from toll like receptor activated, but not rhinovirus infected, primary human airway smooth muscle. Respiratory Research, 14, [127]. https://doi.org/10.1186/1465-9921-14-127 Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). The publication may also be distributed here under the terms of Article 25fa of the Dutch Copyright Act, indicated by the “Taverne” license. More information can be found on the University of Groningen website: https://www.rug.nl/library/open-access/self-archiving-pure/taverne- amendment. Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. -

The Use of Stems in the Selection of International Nonproprietary Names (INN) for Pharmaceutical Substances
WHO/PSM/QSM/2006.3 The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances 2006 Programme on International Nonproprietary Names (INN) Quality Assurance and Safety: Medicines Medicines Policy and Standards The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances FORMER DOCUMENT NUMBER: WHO/PHARM S/NOM 15 © World Health Organization 2006 All rights reserved. Publications of the World Health Organization can be obtained from WHO Press, World Health Organization, 20 Avenue Appia, 1211 Geneva 27, Switzerland (tel.: +41 22 791 3264; fax: +41 22 791 4857; e-mail: [email protected]). Requests for permission to reproduce or translate WHO publications – whether for sale or for noncommercial distribution – should be addressed to WHO Press, at the above address (fax: +41 22 791 4806; e-mail: [email protected]). The designations employed and the presentation of the material in this publication do not imply the expression of any opinion whatsoever on the part of the World Health Organization concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. Dotted lines on maps represent approximate border lines for which there may not yet be full agreement. The mention of specific companies or of certain manufacturers’ products does not imply that they are endorsed or recommended by the World Health Organization in preference to others of a similar nature that are not mentioned. Errors and omissions excepted, the names of proprietary products are distinguished by initial capital letters. -

General Pharmacology
GENERAL PHARMACOLOGY Winners of “Nobel” prize for their contribution to pharmacology Year Name Contribution 1923 Frederick Banting Discovery of insulin John McLeod 1939 Gerhard Domagk Discovery of antibacterial effects of prontosil 1945 Sir Alexander Fleming Discovery of penicillin & its purification Ernst Boris Chain Sir Howard Walter Florey 1952 Selman Abraham Waksman Discovery of streptomycin 1982 Sir John R.Vane Discovery of prostaglandins 1999 Alfred G.Gilman Discovery of G proteins & their role in signal transduction in cells Martin Rodbell 1999 Arvid Carlson Discovery that dopamine is neurotransmitter in the brain whose depletion leads to symptoms of Parkinson’s disease Drug nomenclature: i. Chemical name ii. Non-proprietary name iii. Proprietary (Brand) name Source of drugs: Natural – plant /animal derivatives Synthetic/semisynthetic Plant Part Drug obtained Pilocarpus microphyllus Leaflets Pilocarpine Atropa belladonna Atropine Datura stramonium Physostigma venenosum dried, ripe seed Physostigmine Ephedra vulgaris Ephedrine Digitalis lanata Digoxin Strychnos toxifera Curare group of drugs Chondrodendron tomentosum Cannabis indica (Marijuana) Various parts are used ∆9Tetrahydrocannabinol (THC) Bhang - the dried leaves Ganja - the dried female inflorescence Charas- is the dried resinous extract from the flowering tops & leaves Papaver somniferum, P album Poppy seed pod/ Capsule Natural opiates such as morphine, codeine, thebaine Cinchona bark Quinine Vinca rosea periwinkle plant Vinca alkaloids Podophyllum peltatum the mayapple -

Suplementum 12/1 MAKETA 7/1 2/23/12 12:12 PM Stránka 16
suplementum 12/1 _MAKETA 7/1 2/23/12 12:12 PM Stránka 16 16 ACTA MEDICA MARTINIANA 2012 Suppl. 1 DOI: 10.2478/v10201-011-0023-7 SELECTIVE INHIBITION OF PHOSPHODIESTERASE 7 (PDE7) BY BRL50481 IN HEALTHY AND OVALBUMIN-SENSITIZED GUINEA PIGS Christensen I.1, Miskovicova H. 1, Porvaznik I.1,JoskovaM.1, Mokra D.2, Mokry J.1 1Department of Pharmacology Jessenius Faculty of Medicine, Comenius University, Martin 2Department of Physiology, Jessenius Faculty of Medicine, Comenius University, Martin, Slovak Republic A b stract Phosphodiesterase (PDE) inhibitors may have significant clinical benefit in respiratory diseases associated with inflammation. The aim was to evaluate effects of selective PDE7 inhibitor (BRL50481) on citric acid induced cough, in vivo and in vitro airway smooth muscle reactivity in both healthy and ovalbumin sensitized guinea pigs, as well as its effectiveness in changes of blood cells count. Tested drugs were administered intraperitoneally to male guinea pigs once daily for 7 days either vehicle 10% DMSO (dimethyl sulfoxide) 3 ml/kg (as control) or BRL50481 1 mg/kg. Double chamber whole body plethysmo- graph was used for evaluation of citric acid (0.6 M) induced cough and specific airway resistance. Organ bath method was used for measurement of tracheal and lung tissue strips contractions evoked by cumulative doses (10- 8 – 10-3 mol/L) of acetylcholine (ACH) and histamine (HIS). In healthy guinea pigs we did not observe significant effect of tested drug BRL50481 on in vitro contractions (sim- ilarly to in vivo conditions). The effect on cough was in healthy animals negligible. In ovalbumin-sensitized ani- mals, more pronounced in vitro relaxing effect of BRL50481 in HIS induced contractions was observed with simi- lar results in vivo and no significant change in number of cough efforts. -

New Therapeutic Options in the Management of COPD – Focus on Roflumilast
International Journal of COPD Dovepress open access to scientific and medical research Open Access Full Text Article REVIEW New therapeutic options in the management of COPD – focus on roflumilast Sabina Antonela Antoniu Abstract: In chronic obstructive pulmonary disease (COPD) the inflammation occurring in the University of Medicine and Pharmacy, airways and in other lung tissues is complex and is orchestrated by various mediators including the Pulmonary Disease Division, isoenzyme 4 of the phosphodiesterases family (PDE4), which contributes to bronchoconstriction Pulmonary Disease University and inflammation. Various PDE4 inhibitors have been evaluated as potential therapies in asthma Hospital, Iasi, Romania or COPD but among these only roflumilast have been authorized in Europe to be used in patients with severe COPD as an add-on to the bronchodilator therapy. This review discusses the existing preclinical and clinical data supporting the use of roflumilast for this therapeutic indication and tackles some of the pending issues related to PDE4 in general and to roflumilast in particular. Keywords: COPD, roflumilast, PDE4, efficacy, safety Introduction COPD is an inflammatory disease of the airways related mainly to smoking and char- acterized by airflow limitation, which manifests clinically with dsypnea, cough, and sputum production, symptoms that aggravate disease severity and disease exacerbation. Chronic airways inflammation leads to their progressive obstruction, lung paren- chyma damage, and mucus hypersecretion. Subsequently these result in abnormalities in gas exchange at the pulmonary level and respiratory failure. Currently COPD is classified in 4 stages of severity depending on the degree of airways obstruction measured by FEV1 and with FEV1/FVC% ratio (Table 1). In COPD the therapeutic approach is aimed at treating disease exacerbation when this occurs and at slowing disease progression in the stable state. -
Downloaded from Survive Nursing | Survivenursing.Com V20110426
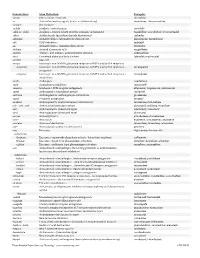
Generic Stem Stem Definition Examples -abine (see -arabine, -citabine) decitabine -ac Anti-inflammatory agents (acetic acid derivatives) bromfenac; dexpemedolac -acetam See -racetam -actide Synthetic corticotropins seractide -adol or -aldol- Analgesics (mixed opiate receptor agonists/ antagonists) tazadolene; spiradolene; levonantradol -adox Antibacterials (quinoline dioxide derivatives) carbadox -afenone Antiarrhythmics (propafenone derivatives) alprafenone; diprafenone -afil PDE5 inhibitors tadalafil -aj- Antiarrhythmics (ajmaline derivatives) lorajmine -aldrate Antacid aluminum salts magaldrate -algron Alpha1 - and alpha2 - adrenoreceptor agonists dabuzalgron -alol Combined alpha and beta blockers labetalol; medroxalol -amivir (see -vir) -ampa Ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) -ampanel Ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) ; becampanel antagonists -ampator Ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) ; forampator modulators -andr- Androgens nandrolone -anib Angiogenesis inhibitors semaxanib -anserin Serotonin 5-HT2 receptor antagonists altanserin; tropanserin; adatanserin -antel Anthelmintics (undefined group) carbantel -antrone Antineoplastics; anthraquinone derivatives pixantrone -apsel P-selectin antagonists torapsel -arabine Antineoplastics (arabinofuranosyl derivatives) fazarabine; fludarabine aril-, -aril, -aril- Antiviral (arildone derivatives) pleconaril; arildone; fosarilate -arit Antirheumatics (lobenzarit type) lobenzarit; clobuzarit -arol