Structural Insights Into Ankyrin Repeat-Containing Proteins and Their Influence in Ubiquitylation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Nedd4 Family of E3 Ubiquitin Ligases: Functional Diversity Within a Common Modular Architecture
Oncogene (2004) 23, 1972–1984 & 2004 Nature Publishing Group All rights reserved 0950-9232/04 $25.00 www.nature.com/onc The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture Robert J Ingham*,1, Gerald Gish1 and Tony Pawson1,2 1Samuel Lunenfeld Research Institute, Mt. Sinai Hospital, Toronto, Ontario, Canada M5G 1X5; 2Department of Molecular and Medical Genetics, University of Toronto, Toronto, Ontario, Canada M5S 1A8 Neuronal precursor cell-expressed developmentally down- Nedd4 was originally identified in 1992, in a screen for regulated 4 (Nedd4)is the prototypical protein in a family genes developmentally downregulated in the early of E3 ubiquitin ligases that have a common domain embryonic mouse central nervous system (Kumar et al., architecture. They are comprised of a catalytic C-terminal 1992), and resembles other regulatory proteins in HECT domain and N-terminal C2 domain and WW containing multiple, distinct modular domains (Pawson domains responsible for cellular localization and substrate and Nash, 2003). Subsequently, related proteins with recognition. These proteins are found throughout eukar- similar structures have been characterized. All contain a yotes and regulate diverse biological processes through the catalytic HECT domain at the C-terminus, and an N- targeted degradation of proteins that generally have a terminal region involved in substrate recognition, that PPxY motif for WW domain recognition, and are found includes a C2 domain and a series of WW domains. in the nucleus and at the plasma membrane. Whereas the Although the common modular architecture of the yeast Saccharomyces cerevisiae uses a single protein, Nedd4 family might suggest a redundant functional role Rsp5p, to carry out these functions, evolution has provided for these proteins, recent work has begun to define higher eukaryotes with several related Nedd4 proteins that specific cellular activities for individual members, and to appear to have specialized roles. -

Ankrd9 Is a Metabolically-Controlled Regulator of Impdh2 Abundance and Macro-Assembly
ANKRD9 IS A METABOLICALLY-CONTROLLED REGULATOR OF IMPDH2 ABUNDANCE AND MACRO-ASSEMBLY by Dawn Hayward A dissertation submitted to The Johns Hopkins University in conformity with the requirements of the degree of Doctor of Philosophy Baltimore, Maryland April 2019 ABSTRACT Members of a large family of Ankyrin Repeat Domains proteins (ANKRD) regulate numerous cellular processes by binding and changing properties of specific protein targets. We show that interactions with a target protein and the functional outcomes can be markedly altered by cells’ metabolic state. ANKRD9 facilitates degradation of inosine monophosphate dehydrogenase 2 (IMPDH2), the rate-limiting enzyme in GTP biosynthesis. Under basal conditions ANKRD9 is largely segregated from the cytosolic IMPDH2 by binding to vesicles. Upon nutrient limitation, ANKRD9 loses association with vesicles and assembles with IMPDH2 into rod-like structures, in which IMPDH2 is stable. Inhibition of IMPDH2 with Ribavirin favors ANKRD9 binding to rods. The IMPDH2/ANKRD9 assembly is reversed by guanosine, which restores association of ANKRD9 with vesicles. The conserved Cys109Cys110 motif in ANKRD9 is required for the vesicles-to-rods transition as well as binding and regulation of IMPDH2. ANKRD9 knockdown increases IMPDH2 levels and prevents formation of IMPDH2 rods upon nutrient limitation. Thus, the status of guanosine pools affects the mode of ANKRD9 action towards IMPDH2. Advisor: Dr. Svetlana Lutsenko, Department of Physiology, Johns Hopkins University School of Medicine Second reader: -

Degradation and Beyond: Control of Androgen Receptor Activity by the Proteasome System
CELLULAR & MOLECULAR BIOLOGY LETTERS Volume 11, (2006) pp 109 – 131 http://www.cmbl.org.pl DOI: 10.2478/s11658-006-0011-9 Received: 06 December 2005 Revised form accepted: 31 January 2006 © 2006 by the University of Wrocław, Poland DEGRADATION AND BEYOND: CONTROL OF ANDROGEN RECEPTOR ACTIVITY BY THE PROTEASOME SYSTEM TOMASZ JAWORSKI Department of Cellular Biochemistry, Nencki Institute of Experimental Biology, Polish Academy of Sciences, Pasteura 3, 02-093 Warsaw, Poland Abstract: The androgen receptor (AR) is a transcription factor belonging to the family of nuclear receptors which mediates the action of androgens in the development of urogenital structures. AR expression is regulated post- * Corresponding author: e-mail: [email protected] Abbreviations used: AATF – apoptosis antagonizing factor; APIS complex – AAA proteins independent of 20S; ARA54 – AR-associated protein 54; ARA70 – AR-associated protein 70; ARE – androgen responsive element; ARNIP – AR N-terminal interacting protein; bFGF – basic fibroblast growth factor; CARM1 – coactivator-associated arginine methyltransferase 1; CHIP – C-terminus Hsp70 interacting protein; DBD – DNA binding domain; E6-AP – E6-associated protein; GRIP-1 – glucocorticoid receptor interacting protein 1; GSK3β – glycogen synthase kinase 3β; HDAC1 – histone deacetylase 1; HECT – homologous to the E6-AP C-terminus; HEK293 – human embryonal kidney cell line; HepG2 – human hepatoma cell line; Hsp90 – heat shock protein 90; IGF-1 – insulin-like growth factor 1; IL-6 – interleukin 6; KLK2 – -

Atnea1-Identification and Characterization of a Novel Plant Nuclear Envelope Associated Protein
Vol. 13(7), pp. 844-856, 12 February, 2014 DOI: 10.5897/AJB2013.13078 ISSN 1684-5315 ©2014 Academic Journals African Journal of Biotechnology http://www.academicjournals.org/AJB Full Length Research Paper AtNEA1-identification and characterization of a novel plant nuclear envelope associated protein Ting Lu Department of Biological and Medical Sciences, Oxford Brookes University, Oxford OX3 0BP, UK. Accepted 29 January, 2014 In animal and yeast cells, a cross nuclear envelope structure linker of nucleoskeleton and cytoskeleton (LINC) is formed by outer nuclear membrane SUN proteins and inner nuclear membrane KASH proteins. However, little information was acquired about plant SUN-KASH structure until they were found in plant SUN proteins in 2010 and KASH proteins in 2012. The SUN-KASH complex alongside with actin in the microfilament cytoskeleton and nucleaoskeleton together shape the main cell skeleton structure and involve in many important biological functions including cell structure stability, cell movement and cell division. In searching of other plant nuclear envelope associated proteins, arabidopsis nuclear envelop associated (AtNEA1) protein 1, a plant nucleoplasmic protein, was identified from biological studies. AtNEA1 was predicted to have a nuclear localisation signal (NLS), two coiled coil domains and one transmembrane (TM) domain. The mutants with the deletion of respective putative domains were observed under confocal microscopy. The subcellular localisation of mutants implied that putative NLS is not essential for AtNEA1 to diffuse through NPC but can strongly increase the efficiency, both coiled coil domains participate in the interaction of AtNEA1 with its unknown INM intrinsic interaction partner, and putative TM appeared to be non-functional. -

View Open Access HPV E6, E6AP and Cervical Cancer Sylvie Beaudenon1 and Jon M Huibregtse*2
BMC Biochemistry BioMed Central Review Open Access HPV E6, E6AP and cervical cancer Sylvie Beaudenon1 and Jon M Huibregtse*2 Address: 1Asuragen, Inc., 2150 Woodward, Austin, TX 78744, USA and 2Molecular Genetics and Microbiology, Institute for Cellular and Molecular Biology, University of Texas at Austin, Austin, TX 78712, USA Email: Jon M Huibregtse* - [email protected] * Corresponding author Published: 21 October 2008 <supplement> <title> <p>Ubiquitin-Proteasome System in Disease Part 2</p> </title> <editor>John Mayer and Rob Layfield</editor> <note>Reviews</note> </supplement> BMC Biochemistry 2008, 9(Suppl 1):S4 doi:10.1186/1471-2091-9-S1-S4 This article is available from: http://www.biomedcentral.com/1471-2091/9/S1/S4 © 2008 Beaudenon and Huibregtse; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Every year, approximately 470,000 new cases of cervical cancer are diagnosed and approximately 230,000 women worldwide die of the disease, with the majority (~80%) of these cases and deaths occurring in developing countries. Human papillomaviruses (HPVs) are the etiological agents in nearly all cases (99.7%) of cervical cancer, and the HPV E6 protein is one of two viral oncoproteins that is expressed in virtually all HPV-positive cancers. E6 hijacks a cellular ubiquitin ligase, E6AP, resulting in the ubiquitylation and degradation of the p53 tumor suppressor, as well as several other cellular proteins. -

Identification of Plant SUN Domain-Interacting Tail Proteins and Analysis of Their Function in Nuclear Positioning
Identification of Plant SUN Domain-Interacting Tail Proteins and Analysis of Their Function in Nuclear Positioning DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of the Ohio State University By Xiao Zhou, M. S. Graduate Program in Plant Cellular and Molecular Biology The Ohio State University 2013 Dissertation Committee: Professor Iris Meier, Advisor Professor Biao Ding Professor Stephen Osmani Professor R. Keith Slotkin Copyright by Xiao Zhou 2013 Abstract The nuclear envelope (NE) is a double membrane system consisting of an inner nuclear membrane (INM) and an outer nuclear membrane (ONM). Studies in opisthokonts revealed that the two membranes are bridged by protein complexes formed by the INM Sad1/UNC-84 (SUN) proteins and the ONM Klarsicht/ANC-1/Syne-1 homology (KASH) proteins. These SUN-KASH NE bridges are usually linkers of the nucleoskeleton to the cytoskeleton (LINC) conserved across eukaryotes. LINC complexes are key players in multiple cellular processes, such as nuclear and chromosomal positioning and nuclear shape determination, which in turn influence the gametogenesis and several aspects of development. Although these cellular processes have long been also known in plants, no KASH proteins are encoded in the plant genomes. I identified WPP domain interacting proteins (WIPs) as the first plant KASH protein analogs. WIPs are plant-specific ONM proteins that redundantly anchor Ran GTPase activating protein (RanGAP) to the NE. Arabidopsis thaliana WIPs (AtWIPs) interact with Arabidopsis thaliana SUN proteins (AtSUNs), which is required for both AtWIP1 and AtRanGAP1 NE localization. In addition, AtWIPs and AtSUNs are necessary for maintaining the elongated nuclear shape of Arabidopsis epidermal cells. -

Repeat Proteins Challenge the Concept of Structural Domains
844 Biochemical Society Transactions (2015) Volume 43, part 5 Repeat proteins challenge the concept of structural domains Rocıo´ Espada*1, R. Gonzalo Parra*1, Manfred J. Sippl†, Thierry Mora‡, Aleksandra M. Walczak§ and Diego U. Ferreiro*2 *Protein Physiology Lab, Dep de Qu´ımica Biologica, ´ Facultad de Ciencias Exactas y Naturales, UBA-CONICET-IQUIBICEN, Buenos Aires, C1430EGA, Argentina †Center of Applied Molecular Engineering, Division of Bioinformatics, Department of Molecular Biology, University of Salzburg, 5020 Salzburg, Austria ‡Laboratoire de physique statistique, CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France §Laboratoire de physique theorique, ´ CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France Abstract Structural domains are believed to be modules within proteins that can fold and function independently. Some proteins show tandem repetitions of apparent modular structure that do not fold independently, but rather co-operate in stabilizing structural forms that comprise several repeat-units. For many natural repeat-proteins, it has been shown that weak energetic links between repeats lead to the breakdown of co-operativity and the appearance of folding sub-domains within an apparently regular repeat array. The quasi-1D architecture of repeat-proteins is crucial in detailing how the local energetic balances can modulate the folding dynamics of these proteins, which can be related to the physiological behaviour of these ubiquitous biological systems. Introduction and between repeats, challenging the concept of structural It was early on noted that many natural proteins typically domain. collapse stretches of amino acid chains into compact units, defining structural domains [1]. These domains typically correlate with biological activities and many modern proteins can be described as composed by novel ‘domain arrange- Definition of the repeat-units ments’ [2]. -

A Structural Guide to Proteins of the NF-Kb Signaling Module
Downloaded from http://cshperspectives.cshlp.org/ on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press A Structural Guide to Proteins of the NF-kB Signaling Module Tom Huxford1 and Gourisankar Ghosh2 1Department of Chemistry and Biochemistry, San Diego State University, 5500 Campanile Drive, San Diego, California 92182-1030 2Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92093-0375 Correspondence: [email protected] The prosurvival transcription factor NF-kB specifically binds promoter DNA to activate target gene expression. NF-kB is regulated through interactions with IkB inhibitor proteins. Active proteolysis of these IkB proteins is, in turn, under the control of the IkB kinase complex (IKK). Together, these three molecules form the NF-kB signaling module. Studies aimed at charac- terizing the molecular mechanisms of NF-kB, IkB, and IKK in terms of their three-dimen- sional structures have lead to a greater understanding of this vital transcription factor system. F-kB is a master transcription factor that from the perspective of their three-dimensional Nresponds to diverse cell stimuli by activat- structures. ing the expression of stress response genes. Multiple signals, including cytokines, growth factors, engagement of the T-cell receptor, and NF-kB bacterial and viral products, induce NF-kB Introduction to NF-kB transcriptional activity (Hayden and Ghosh 2008). A point of convergence for the myriad NF-kB was discovered in the laboratory of of NF-kB inducing signals is the IkB kinase David Baltimore as a nuclear activity with bind- complex (IKK). Active IKK in turn controls ing specificity toward a ten-base-pair DNA transcription factor NF-kB by regulating pro- sequence 50-GGGACTTTCC-30 present within teolysis of the IkB inhibitor protein (Fig. -
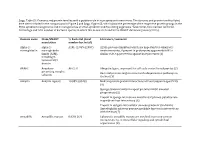
Supp. Table S2: Domains and Protein Families with a Putative Role in Host-Symbiont Interactions
Supp. Table S2: Domains and protein families with a putative role in host-symbiont interactions. The domains and protein families listed here were included in the comparisons in Figure 5 and Supp. Figure S5, which show the percentage of the respective protein groups in the Riftia symbiont metagenome and in metagenomes of other symbiotic and free-living organisms. % bacterial, total number bacterial: Percentage and total number of bacterial species in which this domain is found in the SMART database (January 2019). Domain name Pfam/SMART % bacterial (total Literature/comment annotation number bacterial) Alpha-2- alpha-2- A2M: 42.05% (2057) A2Ms: protease inhibitors which are important for eukaryotic macroglobulin macroglobulin innate immunity, if present in prokaryotes apparently fulfill a family (A2M), similar role, e.g. protection against host proteases (1) including N- terminal MG1 domain ANAPC Anaphase- APC2: 0 Ubiquitin ligase, important for cell cycle control in eukaryotes (2) promoting complex Bacterial proteins might interact with ubiquitination pathways in subunits the host (3) Ankyrin Ankyrin repeats 10.88% (8348) Mediate protein-protein interactions without sequence specificity (4) Sponge symbiont ankyrin-repeat proteins inhibit amoebal phagocytosis (5) Present in sponge microbiome metatranscriptomes, putative role in symbiont-host interactions (6) Present in obligate intracellular amoeba symbiont Candidatus Amoebophilus asiaticus genome, probable function in interactions with the host (7) Armadillo Armadillo repeats 0.83% (67) -

Regulation of P53 Localization and Transcription by the HECT Domain E3 Ligase WWP1
Oncogene (2007) 26, 1477–1483 & 2007 Nature Publishing Group All rights reserved 0950-9232/07 $30.00 www.nature.com/onc SHORT COMMUNICATION Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1 A Laine and Z Ronai Signal Transduction Program, The Burnham Institute for Medical Research, La Jolla, CA, USA As a key cellular regulatory protein p53 is subject to tight 2003; Doran et al., 2004; Shmueli and Oren, 2005; regulation by several E3 ligases. Here, we demonstrate the Harris and Levine, 2005). Common to these ligases is role of HECT domain E3 ligase, WWP1, in regulating p53 their role in limiting p53 stability and activity. In localization and activity. WWP1 associates withp53 and searching for protein ligases with different effects on p53 induces p53 ubiquitylation. Unlike other E3 ligases, activity, we identified the HECT domain E3 ligase WWP1 increases p53 stability; inhibition of WWP1 WWP1, whose interaction with p53 increases its stability expression or expression of a ligase-mutant form results while reducing its transcriptional activity. in decreased p53 expression. WWP1-mediated stabilization WW domain-containing protein 1 (WWP1) was first of p53 is associated withincreased accumulation of p53 in identified as a novelprotein based on its WW modules– cytoplasm witha concomitant decrease in its transcrip- a 35–40 amino-acid (aa) region exhibiting high affinity tional activities. WWP1 effects are independent of Mdm2 towards the PY motif, a proline-rich sequence followed as they are seen in cells lacking Mdm2 expression. by a tyrosine residue (Verdecia et al., 2003). WWP1 Whereas WWP1 limits p53 activity, p53 reduces expres- shares a characteristic domain organization with the E3 sion of WWP1, pointing to a possible feedback loop ligases Nedd4 and Smurfs, which consists of a C2 mechanism. -

Nesprins: from the Nuclear Envelope and Beyond
expert reviews http://www.expertreviews.org/ in molecular medicine Nesprins: from the nuclear envelope and beyond Dipen Rajgor and Catherine M. Shanahan* Nuclear envelope spectrin-repeat proteins (Nesprins), are a novel family of nuclear and cytoskeletal proteins with rapidly expanding roles as intracellular scaffolds and linkers. Originally described as proteins that localise to the nuclear envelope (NE) and establish nuclear-cytoskeletal connections, nesprins have now been found to comprise a diverse spectrum of tissue specific isoforms that localise to multiple sub-cellular compartments. Here, we describe how nesprins are necessary in maintaining cellular architecture by acting as essential scaffolds and linkers at both the NE and other sub-cellular domains. More importantly, we speculate how nesprin mutations may disrupt tissue specific nesprin scaffolds and explain the tissue specific nature of many nesprin-associated diseases, including laminopathies. The eukaryotic cytoplasm contains three major composed of three α-helical bundles with a left- types of cytoskeletal filaments: Filamentous- handed twist, and its primary function is to actin (F-actin), microtubules (MTs) and provide docking sites for proteins and other intermediate filaments (IFs). These components higher order complexes (Refs 3, 4). Although are organised in a manner that provides the cell most SR proteins contain CHDs, some possess with an internal framework fundamental for motifs which can interact with other cytoskeletal many processes, such as controlling cellular components, allowing linkage of SR-associated shape, polarity, adhesion and migration, complexes to filamentous structures other than cytokinesis, inter- and intracellular F-actin. In addition, these motifs allow cross- Nesprins: from the nuclear envelope and beyond communication and trafficking of organelles, linking between different filaments and dynamic vesicles, proteins and RNA (Refs 1, 2). -

The Nuclear Envelope: Target and Mediator of the Apoptotic Process Liora Lindenboim1, Hila Zohar1,Howardj.Worman2 and Reuven Stein1
Lindenboim et al. Cell Death Discovery (2020) 6:29 https://doi.org/10.1038/s41420-020-0256-5 Cell Death Discovery REVIEW ARTICLE Open Access The nuclear envelope: target and mediator of the apoptotic process Liora Lindenboim1, Hila Zohar1,HowardJ.Worman2 and Reuven Stein1 Abstract Apoptosis is characterized by the destruction of essential cell organelles, including the cell nucleus. The nuclear envelope (NE) separates the nuclear interior from the cytosol. During apoptosis, the apoptotic machinery, in particular caspases, increases NE permeability by cleaving its proteins, such as those of the nuclear pore complex (NPC) and the nuclear lamina. This in turns leads to passive diffusion of cytosolic apoptogenic proteins, such as caspases and nucleases, through NPCs into the nucleus and the subsequent breakdown of the NE and destruction of the nucleus. However, NE leakiness at early stages of the apoptotic process can also occur in a caspase-independent manner, where Bax, by a non-canonical action, promotes transient and repetitive localized generation and subsequent rupture of nuclear protein-filled nuclear bubbles. This NE rupture leads to discharge of apoptogenic nuclear proteins from the nucleus to the cytosol, a process that can contribute to the death process. Therefore, the NE may play a role as mediator of cell death at early stages of apoptosis. The NE can also serve as a platform for assembly of complexes that regulate the death process. Thus, the NE should be viewed as both a mediator of the cell death process and a target. 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; Facts redistribution of the nuclear proteins to the cytosol that might subsequently act as amplifiers of the ● The NE is an important target of the apoptotic apoptotic process.