Long Noncoding Rnas Are Critical Regulators of Pancreatic Islet Development and Function
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Expression of NOD2, NLRP3 and NLRC5 and Renal Injury in Anti-Neutrophil Cytoplasmic Antibody-Associated Vasculitis
Wang et al. J Transl Med (2019) 17:197 https://doi.org/10.1186/s12967-019-1949-5 Journal of Translational Medicine RESEARCH Open Access The expression of NOD2, NLRP3 and NLRC5 and renal injury in anti-neutrophil cytoplasmic antibody-associated vasculitis Luo‑Yi Wang1,2,3, Xiao‑Jing Sun1,2,3, Min Chen1,2,3* and Ming‑Hui Zhao1,2,3,4 Abstract Background: Nucleotide‑binding oligomerization domain (NOD)‑like receptors (NLRs) are intracellular sensors of pathogens and molecules from damaged cells to regulate the infammatory response in the innate immune system. Emerging evidences suggested a potential role of NLRs in anti‑neutrophil cytoplasmic antibody (ANCA)‑associated vasculitis (AAV). This study aimed to investigate the expression of nucleotide‑binding oligomerization domain con‑ taining protein 2 (NOD2), NOD‑like receptor family pyrin domain containing 3 (NLRP3) and NOD‑like receptor family CARD domain containing 5 (NLRC5) in kidneys of AAV patients, and further explored their associations with clinical and pathological parameters. Methods: Thirty‑four AAV patients in active stage were recruited. Their renal specimens were processed with immu‑ nohistochemistry to assess the expression of three NLRs, and with double immunofuorescence to detect NLRs on intrinsic and infltrating cells. Analysis of gene expression was also adopted in cultured human podocytes. The associa‑ tions between expression of NLRs and clinicopathological parameters were analyzed. Results: The expression of NOD2, NLRP3 and NLRC5 was signifcantly higher in kidneys from AAV patients than those from normal controls, minimal change disease or class IV lupus nephritis. These NLRs co‑localized with podocytes and infltrating infammatory cells. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Examination of the Transcription Factors Acting in Bone Marrow
THESIS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY (PHD) Examination of the transcription factors acting in bone marrow derived macrophages by Gergely Nagy Supervisor: Dr. Endre Barta UNIVERSITY OF DEBRECEN DOCTORAL SCHOOL OF MOLECULAR CELL AND IMMUNE BIOLOGY DEBRECEN, 2016 Table of contents Table of contents ........................................................................................................................ 2 1. Introduction ............................................................................................................................ 5 1.1. Transcriptional regulation ................................................................................................... 5 1.1.1. Transcriptional initiation .................................................................................................. 5 1.1.2. Co-regulators and histone modifications .......................................................................... 8 1.2. Promoter and enhancer sequences guiding transcription factors ...................................... 11 1.2.1. General transcription factors .......................................................................................... 11 1.2.2. The ETS superfamily ..................................................................................................... 17 1.2.3. The AP-1 and CREB proteins ........................................................................................ 20 1.2.4. Other promoter specific transcription factor families ................................................... -

NFX1, Its Isoforms and Roles in Biology, Disease and Cancer
biology Review NFX1, Its Isoforms and Roles in Biology, Disease and Cancer Sreenivasulu Chintala and Rachel A. Katzenellenbogen * Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN 46202, USA; [email protected] * Correspondence: [email protected] Simple Summary: The NFX1 gene, and its gene products, were identified over 30 years ago. Since then, the literature on NFX1 homologs and NFX1 itself has grown. In this review, we summarize the studies to-date on the NFX1 gene and its proteins across species and in humans, describing their role in gene regulation, embryonic development, cellular growth and differentiation, exogenous stress tolerance and metabolism, and an organism’s immune response. We also highlight the roles NFX1 has in human disease and in cancer, with a strong focus on its collaborative role with high-risk human papillomavirus infections that cause cervical and head and neck cancers. We believe this is the first comprehensive review of NFX1 and its functional significance in organisms ranging from yeast to human. Abstract: In 1989, two NFX1 protein products were identified as nuclear proteins with the ability to bind to X-box cis-elements. Since that publication, the NFX1 gene and its homologs have been identified, from yeast to humans. This review article summarizes what is known about the NFX1 gene across species. We describe the gene and protein motifs of NFX1 homologs and their functions in cellular biology, then turn to NFX1 in human biology and disease development. In that, we focus on more recent literature about NFX1 and its two splice variants protein products (NFX1-91 and NFX1- Citation: Chintala, S.; 123) that are expressed in epithelial cells. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Distinguishing Pleiotropy from Linked QTL Between Milk Production Traits
Cai et al. Genet Sel Evol (2020) 52:19 https://doi.org/10.1186/s12711-020-00538-6 Genetics Selection Evolution RESEARCH ARTICLE Open Access Distinguishing pleiotropy from linked QTL between milk production traits and mastitis resistance in Nordic Holstein cattle Zexi Cai1*†, Magdalena Dusza2†, Bernt Guldbrandtsen1, Mogens Sandø Lund1 and Goutam Sahana1 Abstract Background: Production and health traits are central in cattle breeding. Advances in next-generation sequencing technologies and genotype imputation have increased the resolution of gene mapping based on genome-wide association studies (GWAS). Thus, numerous candidate genes that afect milk yield, milk composition, and mastitis resistance in dairy cattle are reported in the literature. Efect-bearing variants often afect multiple traits. Because the detection of overlapping quantitative trait loci (QTL) regions from single-trait GWAS is too inaccurate and subjective, multi-trait analysis is a better approach to detect pleiotropic efects of variants in candidate genes. However, large sample sizes are required to achieve sufcient power. Multi-trait meta-analysis is one approach to deal with this prob- lem. Thus, we performed two multi-trait meta-analyses, one for three milk production traits (milk yield, protein yield and fat yield), and one for milk yield and mastitis resistance. Results: For highly correlated traits, the power to detect pleiotropy was increased by multi-trait meta-analysis com- pared with the subjective assessment of overlapping of single-trait QTL confdence intervals. Pleiotropic efects of lead single nucleotide polymorphisms (SNPs) that were detected from the multi-trait meta-analysis were confrmed by bivariate association analysis. The previously reported pleiotropic efects of variants within the DGAT1 and MGST1 genes on three milk production traits, and pleiotropic efects of variants in GHR on milk yield and fat yield were con- frmed. -

Transcriptional Control of Tissue-Resident Memory T Cell Generation
Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2019 © 2019 Filip Cvetkovski All rights reserved ABSTRACT Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Tissue-resident memory T cells (TRM) are a non-circulating subset of memory that are maintained at sites of pathogen entry and mediate optimal protection against reinfection. Lung TRM can be generated in response to respiratory infection or vaccination, however, the molecular pathways involved in CD4+TRM establishment have not been defined. Here, we performed transcriptional profiling of influenza-specific lung CD4+TRM following influenza infection to identify pathways implicated in CD4+TRM generation and homeostasis. Lung CD4+TRM displayed a unique transcriptional profile distinct from spleen memory, including up-regulation of a gene network induced by the transcription factor IRF4, a known regulator of effector T cell differentiation. In addition, the gene expression profile of lung CD4+TRM was enriched in gene sets previously described in tissue-resident regulatory T cells. Up-regulation of immunomodulatory molecules such as CTLA-4, PD-1, and ICOS, suggested a potential regulatory role for CD4+TRM in tissues. Using loss-of-function genetic experiments in mice, we demonstrate that IRF4 is required for the generation of lung-localized pathogen-specific effector CD4+T cells during acute influenza infection. Influenza-specific IRF4−/− T cells failed to fully express CD44, and maintained high levels of CD62L compared to wild type, suggesting a defect in complete differentiation into lung-tropic effector T cells. -

Striking the Right Balance in Anti-Neutrophil Cytoplasmic Antibody-Associated Vasculitis
Striking the Right Balance in Anti-neutrophil Cytoplasmic Antibody-Associated Vasculitis This symposium took place on 4th June 2021, as part of the European Alliance of Associations for Rheumatology (EULAR) virtual congress Speakers: Benjamin Terrier,¹ Joanna Robson,² Bernhard Hellmich³ 1. University of Paris and Hôpital Cochin, France 2. University of the West of England and Bristol Royal Infirmary, UK 3. University of Tübingen, Germany Disclosure: Terrier has been an advisory board member and/or received consulting fees/ travel expenses from AstraZeneca, Chugai, Grifols, GlaxoSmithKline, Janssen, LFB, Octapharma, Roche, and Vifor Pharma. Robson has received speaker’s fees from Roche and Vifor Pharma; and research support from Vifor Pharma. Hellmich has been an investigator in clinical trials for Ab2Bio, AbbVie, AstraZeneca, Bristol- Myers Squibb, Chemocentrix, GlaxoSmithKline, InflaRx, Kiniksa, Nippon Kayaku, Novartis, Roche, and Sanofi. He has acted as a consultant, advisory board member, and/or lecturer for AbbVie, Bristol-Myers Squibb, Boehringer Ingelheim, Chugai, GlaxoSmithKline, InflaRx, Novartis, Pfizer, Roche, and Vifor Pharma. He is also a member of the Guideline Committees for European Alliance of Associations for Rheumatology (EULAR) and the German Society of Rheumatology (DGRh). Acknowledgements: Writing assistance was provided by Helen Boreham. Support: The publication of this article was funded by Vifor Pharma. The views and opinions expressed are those of the presenters. Content was reviewed by Vifor Pharma for medical accuracy. Citation: Rheumatol. 2021;8[1]:43-50. Meeting Summary Anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV) causes irreversible short- and long-term damage to vital organs, particularly the kidneys and lungs. Current standard of care (SOC) for AAV, of which glucocorticoids (GC) are a lynchpin, has a number of important limitations: responses to therapy are variable, some patients fail to achieve and sustain remission, and treatment related adverse events (AE) are common. -
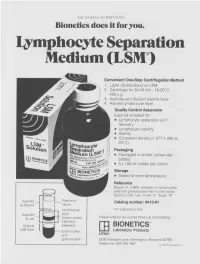
Lymphocyte Separation Medium (LSM
THE JOURNAL OF IMMUNOLOGY Bionetics does it for you. Lymphocyte Separation Medium (LSM wenient One-Step Centrifugation Method _ayer diluted blood on LSM. 2,entrifuge for 30-40 min., 18-20°C, ~.00 x g. ~,spirate and discard plasma layer -larvest lymphocyte layer. Quality Control Assurance Each lot is tested for: • Lymphocyte separation and recovery. • Lymphocyte viability. • Sterility. • Consistent density (1.077-1.080 at 20°C). Packaging • Packaged in amber, screw-cap bottles. • 5 x 100 ml bottles per carton. Storage. • Stored at room temperature. Reference Boyum, A. (1968): Isolation of mononuclear cells and granulocytes from human blood. Scand J. Clin. Lab. Invest. 21, Suppl. 97. Aspirate I IC[OI I IC~ LJI Catalog number: 8410-01 & discard serum Lymphocyte For Laboratory Use Aspirate layer & use (mononuclear Please write for our current Price List and Catalog. cells and Original platelets) ITi BIONETICS° LSM layer Erythrocytes Laboratory Products and Litton granulocytes 5516 Nicholson Lane, Kensington, Maryland 20795 Telephone: (301) 881-1557 1979 Litton Bionetics, tnc Get the most out of your high quality cytotoxic antibodies with LOW-TOX-M RABBIT COMPLEMENT LOW TOXICITY HIGH ACTIVITY Presentation: CL 3051 5 x 1 ml, lyophilized $30.00 When it comes to COMPLEMENT... come to CEDARLANE Direct orders or inquiries to: UNITED STATES: WORLDWIDE EXCEPT U.S. ,4 C~L CEDARLANE ACCURATE CHEMICAL & LABORATO RI ES SCIENTIFIC CORPORATION LIMITED 5516-8TH LINE, R.R. 2 28 TEC STREET, HICKSVtLLE, N.Y. 11801 HORNBY, ONTARIO, CANADA LOP 1E0 Telephone -

Análise Integrativa De Perfis Transcricionais De Pacientes Com
UNIVERSIDADE DE SÃO PAULO FACULDADE DE MEDICINA DE RIBEIRÃO PRETO PROGRAMA DE PÓS-GRADUAÇÃO EM GENÉTICA ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Ribeirão Preto – 2012 ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas Tese apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo para obtenção do título de Doutor em Ciências. Área de Concentração: Genética Orientador: Prof. Dr. Eduardo Antonio Donadi Co-orientador: Prof. Dr. Geraldo A. S. Passos Ribeirão Preto – 2012 AUTORIZO A REPRODUÇÃO E DIVULGAÇÃO TOTAL OU PARCIAL DESTE TRABALHO, POR QUALQUER MEIO CONVENCIONAL OU ELETRÔNICO, PARA FINS DE ESTUDO E PESQUISA, DESDE QUE CITADA A FONTE. FICHA CATALOGRÁFICA Evangelista, Adriane Feijó Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. Ribeirão Preto, 2012 192p. Tese de Doutorado apresentada à Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo. Área de Concentração: Genética. Orientador: Donadi, Eduardo Antonio Co-orientador: Passos, Geraldo A. 1. Expressão gênica – microarrays 2. Análise bioinformática por module maps 3. Diabetes mellitus tipo 1 4. Diabetes mellitus tipo 2 5. Diabetes mellitus gestacional FOLHA DE APROVAÇÃO ADRIANE FEIJÓ EVANGELISTA Análise integrativa de perfis transcricionais de pacientes com diabetes mellitus tipo 1, tipo 2 e gestacional, comparando-os com manifestações demográficas, clínicas, laboratoriais, fisiopatológicas e terapêuticas. -

Supporting Information
Supporting Information Pouryahya et al. SI Text Table S1 presents genes with the highest absolute value of Ricci curvature. We expect these genes to have significant contribution to the network’s robustness. Notably, the top two genes are TP53 (tumor protein 53) and YWHAG gene. TP53, also known as p53, it is a well known tumor suppressor gene known as the "guardian of the genome“ given the essential role it plays in genetic stability and prevention of cancer formation (1, 2). Mutations in this gene play a role in all stages of malignant transformation including tumor initiation, promotion, aggressiveness, and metastasis (3). Mutations of this gene are present in more than 50% of human cancers, making it the most common genetic event in human cancer (4, 5). Namely, p53 mutations play roles in leukemia, breast cancer, CNS cancers, and lung cancers, among many others (6–9). The YWHAG gene encodes the 14-3-3 protein gamma, a member of the 14-3-3 family proteins which are involved in many biological processes including signal transduction regulation, cell cycle pro- gression, apoptosis, cell adhesion and migration (10, 11). Notably, increased expression of 14-3-3 family proteins, including protein gamma, have been observed in a number of human cancers including lung and colorectal cancers, among others, suggesting a potential role as tumor oncogenes (12, 13). Furthermore, there is evidence that loss Fig. S1. The histogram of scalar Ricci curvature of 8240 genes. Most of the genes have negative scalar Ricci curvature (75%). TP53 and YWHAG have notably low of p53 function may result in upregulation of 14-3-3γ in lung cancer Ricci curvatures. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int.