Primepcr™Assay Validation Report
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genome-Wide Analysis of 5-Hmc in the Peripheral Blood of Systemic Lupus Erythematosus Patients Using an Hmedip-Chip
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 35: 1467-1479, 2015 Genome-wide analysis of 5-hmC in the peripheral blood of systemic lupus erythematosus patients using an hMeDIP-chip WEIGUO SUI1*, QIUPEI TAN1*, MING YANG1, QIANG YAN1, HUA LIN1, MINGLIN OU1, WEN XUE1, JIEJING CHEN1, TONGXIANG ZOU1, HUANYUN JING1, LI GUO1, CUIHUI CAO1, YUFENG SUN1, ZHENZHEN CUI1 and YONG DAI2 1Guangxi Key Laboratory of Metabolic Diseases Research, Central Laboratory of Guilin 181st Hospital, Guilin, Guangxi 541002; 2Clinical Medical Research Center, the Second Clinical Medical College of Jinan University (Shenzhen People's Hospital), Shenzhen, Guangdong 518020, P.R. China Received July 9, 2014; Accepted February 27, 2015 DOI: 10.3892/ijmm.2015.2149 Abstract. Systemic lupus erythematosus (SLE) is a chronic, Introduction potentially fatal systemic autoimmune disease characterized by the production of autoantibodies against a wide range Systemic lupus erythematosus (SLE) is a typical systemic auto- of self-antigens. To investigate the role of the 5-hmC DNA immune disease, involving diffuse connective tissues (1) and modification with regard to the onset of SLE, we compared is characterized by immune inflammation. SLE has a complex the levels 5-hmC between SLE patients and normal controls. pathogenesis (2), involving genetic, immunologic and envi- Whole blood was obtained from patients, and genomic DNA ronmental factors. Thus, it may result in damage to multiple was extracted. Using the hMeDIP-chip analysis and valida- tissues and organs, especially the kidneys (3). SLE arises from tion by quantitative RT-PCR (RT-qPCR), we identified the a combination of heritable and environmental influences. differentially hydroxymethylated regions that are associated Epigenetics, the study of changes in gene expression with SLE. -

Anti-PPAP2A (Internal) Polyclonal Antibody (CABT- B799) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Anti-PPAP2A (Internal) polyclonal antibody (CABT- B799) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Target PPAP2A Immunogen Synthetic peptide Isotype IgG Source/Host Rabbit Species Reactivity Human, Mouse, Zebrafish Purification Protein A Purified Total IgG Conjugate Unconjugated Applications IHC, WB Format Liquid Concentration 1 mg/ml (Please refer to the vial label for the specific concentration) Size 100 μg Buffer Phosphate buffered saline (PBS) Preservative None Storage Store at 4°C for short-term use. For longer periods of storage, aliquot and store at -20°C. Avoid repeat freeze-thaw cycles. BACKGROUND Introduction PPAP2A is a member of the phosphatidic acid phosphatase (PAP) family. PAPs convert phosphatidic acid to diacylglycerol, and function in de novo synthesis of glycerolipids as well as in receptor-activated signal transduction mediated by phospholipase D. This protein is an integral membrane glycoprotein, and has been shown to be a surface enzyme that plays an active role in the hydrolysis and uptake of lipids from extracellular space. The expression of PPAP2A is found to be regulated by androgen in a prostatic adenocarcinoma cell line.The protein encoded by this 45-1 Ramsey Road, Shirley, NY 11967, USA Email: [email protected] Tel: 1-631-624-4882 Fax: 1-631-938-8221 1 © Creative Diagnostics All Rights Reserved gene is a member of the phosphatidic acid phosphatase (PAP) family. PAPs convert phosphatidic acid to diacylglycerol, and function in de novo synthesis of glycerolipids as well as in receptor-activated signal transduction mediated by phospholipase D. This protein is an integral membrane glycoprotein, and has been shown to be a surface enzyme that plays an active role in the hydrolysis and uptake of lipids from extracellular space. -

PPAP2A (PLPP1) (NM 176895) Human Tagged ORF Clone Lentiviral Particle – RC223188L4V | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC223188L4V PPAP2A (PLPP1) (NM_176895) Human Tagged ORF Clone Lentiviral Particle Product data: Product Type: Lentiviral Particles Product Name: PPAP2A (PLPP1) (NM_176895) Human Tagged ORF Clone Lentiviral Particle Symbol: PLPP1 Synonyms: LLP1a; LPP1; PAP-2a; PAP2; PPAP2A Vector: pLenti-C-mGFP-P2A-Puro (PS100093) ACCN: NM_176895 ORF Size: 855 bp ORF Nucleotide The ORF insert of this clone is exactly the same as(RC223188). Sequence: OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_176895.1 RefSeq Size: 1641 bp RefSeq ORF: 858 bp Locus ID: 8611 UniProt ID: O14494, A0A024QZU7 Protein Families: Druggable Genome, Transmembrane Protein Pathways: Ether lipid metabolism, Fc gamma R-mediated phagocytosis, Glycerolipid metabolism, Glycerophospholipid metabolism, Metabolic pathways, Sphingolipid metabolism MW: 32 kDa This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 PPAP2A (PLPP1) (NM_176895) Human Tagged ORF Clone Lentiviral Particle – RC223188L4V Gene Summary: The protein encoded by this gene is a member of the phosphatidic acid phosphatase (PAP) family. -
Stimulated Phospholipid Synthesis Is Key for Hepatitis B Virus Replications
www.nature.com/scientificreports OPEN Stimulated phospholipid synthesis is key for hepatitis B virus replications Received: 8 March 2019 Qingxia Huang 1,2, Hehua Lei1, Laifeng Ding1,2 & Yulan Wang 3 Accepted: 19 August 2019 Chronic hepatitis B Virus (HBV) infection has high morbidity, high pathogenicity and unclear Published: xx xx xxxx pathogenesis. To elucidate the relationship between HBV replication and host phospholipid metabolites, we measured 10 classes of phospholipids in serum of HBV infected patients and cells using ultra performance liquid chromatograph-triple quadruple mass spectrometry. We found that the levels of phosphatidylcholine (PC), phosphatidylethanolamine, and lyso-phosphatidic acid were increased in HBsAg (+) serum of infected patients compared with HBsAg (−), while phosphatidylserine, phosphatidylglycerol, phosphatidylinositol, and sphingomyelin were decreased, which were confrmed in an HBV infected HepG2.2.15 cell line. We further evaluated the enzyme levels of PC pathways and found that PCYT1A and LPP1 for PC synthesis were up-regulated after HBV infection. Moreover, HBV replication was inhibited when PCYT1A and LPP1 were inhibited. These results indicated that the PC synthesis in HBV infected host are regulated by PCYT1A and LPP1, which suggests that PCYT1A, LPP1 could be new potential targets for HBV treatment. Hepatitis B Virus (HBV) is a double stranded DNA virus and belongs to hepadnavirdae family. HBV infection causes a severe liver infectious disease and has become a global problem afecting human health. In 2015, WHO estimated that approximately 257 million people, equivalent to 3.5% of the population, were infected with chronic HBV worldwide, resulting in 887, 000 HBV-related deaths annually1. Many liver conditions are associated with HBV infection, which include acute hepatitis, chronic hepatitis B (CHB), cirrhosis and hepatocellular carcinoma (HCC)2,3. -
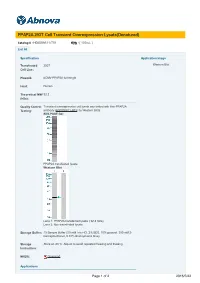
PPAP2A 293T Cell Transient Overexpression Lysate(Denatured)
PPAP2A 293T Cell Transient Overexpression Lysate(Denatured) Catalog # : H00008611-T01 規格 : [ 100 uL ] List All Specification Application Image Transfected 293T Western Blot Cell Line: Plasmid: pCMV-PPAP2A full-length Host: Human Theoretical MW 32.2 (kDa): Quality Control Transient overexpression cell lysate was tested with Anti-PPAP2A Testing: antibody (H00008611-B01) by Western Blots. SDS-PAGE Gel PPAP2A transfected lysate. Western Blot Lane 1: PPAP2A transfected lysate ( 32.2 KDa) Lane 2: Non-transfected lysate. Storage Buffer: 1X Sample Buffer (50 mM Tris-HCl, 2% SDS, 10% glycerol, 300 mM 2- mercaptoethanol, 0.01% Bromophenol blue) Storage Store at -80°C. Aliquot to avoid repeated freezing and thawing. Instruction: MSDS: Download Applications Page 1 of 2 2016/5/22 Western Blot Gene Information Entrez GeneID: 8611 GeneBank NM_003711 Accession#: Protein NP_003702.2 Accession#: Gene Name: PPAP2A Gene Alias: LLP1a,LPP1,PAP-2a,PAP2,PAP2a2,PAP2alpha2,PAPalpha1 Gene phosphatidic acid phosphatase type 2A Description: Omim ID: 607124 Gene Ontology: Hyperlink Gene Summary: The protein encoded by this gene is a member of the phosphatidic acid phosphatase (PAP) family. PAPs convert phosphatidic acid to diacylglycerol, and function in de novo synthesis of glycerolipids as well as in receptor-activated signal transduction mediated by phospholipase D. This protein is an integral membrane glycoprotein, and has been shown to be a surface enzyme that plays an active role in the hydrolysis and uptake of lipids from extracellular space. The expression of this gene is found to be regulated by androgen in a prostatic adenocarcinoma cell line. At least two alternatively spliced transcript variants encoding distinct isoforms have been described. -

CREB-Dependent Transcription in Astrocytes: Signalling Pathways, Gene Profiles and Neuroprotective Role in Brain Injury
CREB-dependent transcription in astrocytes: signalling pathways, gene profiles and neuroprotective role in brain injury. Tesis doctoral Luis Pardo Fernández Bellaterra, Septiembre 2015 Instituto de Neurociencias Departamento de Bioquímica i Biologia Molecular Unidad de Bioquímica y Biologia Molecular Facultad de Medicina CREB-dependent transcription in astrocytes: signalling pathways, gene profiles and neuroprotective role in brain injury. Memoria del trabajo experimental para optar al grado de doctor, correspondiente al Programa de Doctorado en Neurociencias del Instituto de Neurociencias de la Universidad Autónoma de Barcelona, llevado a cabo por Luis Pardo Fernández bajo la dirección de la Dra. Elena Galea Rodríguez de Velasco y la Dra. Roser Masgrau Juanola, en el Instituto de Neurociencias de la Universidad Autónoma de Barcelona. Doctorando Directoras de tesis Luis Pardo Fernández Dra. Elena Galea Dra. Roser Masgrau In memoriam María Dolores Álvarez Durán Abuela, eres la culpable de que haya decidido recorrer el camino de la ciencia. Que estas líneas ayuden a conservar tu recuerdo. A mis padres y hermanos, A Meri INDEX I Summary 1 II Introduction 3 1 Astrocytes: physiology and pathology 5 1.1 Anatomical organization 6 1.2 Origins and heterogeneity 6 1.3 Astrocyte functions 8 1.3.1 Developmental functions 8 1.3.2 Neurovascular functions 9 1.3.3 Metabolic support 11 1.3.4 Homeostatic functions 13 1.3.5 Antioxidant functions 15 1.3.6 Signalling functions 15 1.4 Astrocytes in brain pathology 20 1.5 Reactive astrogliosis 22 2 The transcription -

Chemico-Biological Interactions 196 (2012) 89–95
Chemico-Biological Interactions 196 (2012) 89–95 Contents lists available at ScienceDirect Chemico-Biological Interactions journal homepage: www.elsevier.com/locate/chembioint Exposure to sodium tungstate and Respiratory Syncytial Virus results in hematological/immunological disease in C57BL/6J mice ⇑ Cynthia D. Fastje a, , Kevin Harper a, Chad Terry a, Paul R. Sheppard b, Mark L. Witten a,1 a Steele Children’s Research Center, PO Box 245073, University of Arizona, Tucson, AZ 85724-5073, USA b Laboratory of Tree-Ring Research, PO Box 210058, University of Arizona, Tucson, AZ 85721-0058, USA article info abstract Article history: The etiology of childhood leukemia is not known. Strong evidence indicates that precursor B-cell Acute Available online 1 May 2011 Lymphoblastic Leukemia (Pre-B ALL) is a genetic disease originating in utero. Environmental exposures in two concurrent, childhood leukemia clusters have been profiled and compared with geographically Keywords: similar control communities. The unique exposures, shared in common by the leukemia clusters, have Tungsten been modeled in C57BL/6 mice utilizing prenatal exposures. This previous investigation has suggested Respiratory Syncytial Virus in utero exposure to sodium tungstate (Na2WO4) may result in hematological/immunological disease Childhood leukemia through genes associated with viral defense. The working hypothesis is (1) in addition to spontaneously and/or chemically generated genetic lesions forming pre-leukemic clones, in utero exposure to Na2WO4 increases genetic susceptibility to viral influence(s); (2) postnatal exposure to a virus possessing the 1FXXKXFXXA/V9 peptide motif will cause an unnatural immune response encouraging proliferation in the B-cell precursor compartment. This study reports the results of exposing C57BL/6J mice to Na2WO4 3 in utero via water (15 ppm, ad libetum) and inhalation (mean concentration PM5 3.33 mg/m ) and to Respiratory Syncytial Virus (RSV) within 2 weeks of weaning. -

Expression-Based GWAS Identifies Variants, Gene Interactions and Key
www.nature.com/scientificreports OPEN Expression-based GWAS identifies variants, gene interactions and key regulators affecting intramuscular Received: 12 February 2016 Accepted: 26 July 2016 fatty acid content and composition Published: 18 August 2016 in porcine meat Anna Puig-Oliveras1,2, Manuel Revilla1,2, Anna Castelló1,2, Ana I. Fernández3, Josep M. Folch1,2 & Maria Ballester1,2,4 The aim of this work is to better understand the genetic mechanisms determining two complex traits affecting porcine meat quality: intramuscular fat (IMF) content and its fatty acid (FA) composition. With this purpose, expression Genome-Wide Association Study (eGWAS) of 45 lipid-related genes associated with meat quality traits in swine muscle (Longissimus dorsi) of 114 Iberian × Landrace backcross animals was performed. The eGWAS identified 241 SNPs associated with 11 genes:ACSM5, CROT, FABP3, FOS, HIF1AN, IGF2, MGLL, NCOA1, PIK3R1, PLA2G12A and PPARA. Three expression Quantitative Trait Loci (eQTLs) for IGF2, ACSM5 and MGLL were identified, showingcis -acting effects, whereas 16 eQTLs had trans regulatory effects. A polymorphism in theACSM5 promoter region associated with its expression was identified. In addition, strong candidate genes regulatingACSM5, FOS, PPARA, PIK3R1, PLA2G12A and HIF1AN gene expression were also seen. Notably, the analysis highlighted the NR3C1 transcription factor as a strong candidate gene involved in the regulation of the 45 genes analysed. Finally, the IGF2, MGLL, MC2R, ARHGAP6, and NR3C1 genes were identified as potential regulators co-localizing within QTLs for fatness and growth traits in the IBMAP population. The results obtained increase our knowledge in the functional regulatory mechanisms involved in these complex traits. Pork-meat cuts and their derived products are paid according to the lean percentage in pork carcasses and meat quality, since they determine better acceptance for consumers1. -

PPAP2A Antibody (C-Term) Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap21024c
苏州工业园区双圩路9号1幢 邮 编 : 215000 电 话 : 0512-88856768 PPAP2A Antibody (C-term) Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP21024c Specification PPAP2A Antibody (C-term) - Product info Application WB Primary Accession O14494 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Clone Names RB51855 Calculated MW 32156 PPAP2A Antibody (C-term) - Additional info Gene ID 8611 Other Names Lipid phosphate phosphohydrolase 1, PAP2-alpha, Phosphatidate phosphohydrolase type 2a, Phosphatidic acid phosphatase 2a, PAP-2a, PAP2a, PPAP2A, LPP1 Western blot analysis of lysate from Target/Specificity Jurkat cell line, using PPAP2A Antibody This PPAP2A antibody is generated from a rabbit immunized (C-term)(Cat. #AP21024c). AP21024c was with a KLH conjugated synthetic peptide between 270-304 diluted at 1:1000. A goat anti-rabbit IgG amino acids from the C-terminal region of human PPAP2A. H&L(HRP) at 1:10000 dilution was used as the secondary antibody. Lysate at 20ug. Dilution WB~~1:1000 Format Purified polyclonal antibody supplied in PBS with 0.09% (W/V) sodium azide. This antibody is purified through a protein A column, followed by peptide affinity purification. Storage Maintain refrigerated at 2-8°C for up to 2 weeks. For long term storage store at -20°C in small aliquots to prevent freeze-thaw cycles. Precautions PPAP2A Antibody (C-term) is for research use only and not for use in diagnostic or therapeutic procedures. PPAP2A Antibody (C-term) - Protein Information Western blot analysis of lysate from Name PLPP1 (HGNC:9228) human kidney tissue lysate, using PPAP2A Antibody (C-term)(Cat. Synonyms LPP1, PPAP2A #AP21024c). AP21024c was diluted at 1:1000. -

Analyzing the Role of Genetics and Genomics in Cardiovascular Disease
IT'S COMPLICATED: ANALYZING THE ROLE OF GENETICS AND GENOMICS IN CARDIOVASCULAR DISEASE by JEFFREY HSU Submitted in partial fulfillment of the requirements For the degree of Doctor of Philosophy Dissertation Adviser: Dr. Jonathan D Smith Department of Molecular Medicine CASE WESTERN RESERVE UNIVERSITY January 2014 CASE WESTERN RESERVE UNIVERSITY SCHOOL OF GRADUATE STUDIES We hereby approve the thesis/dissertation of: Jeffrey Hsu candidate for the Doctor of Philosophy degree*. Thomas LaFramboise Thesis Committee Chair Mina Chung David Van Wagoner David Serre Jonathan D Smith Date: 5/24/2013 We also certify that written approval has been obtained for any proprietary material contained therein. i Contents Abstract 1 Acknowledgments 3 1 Introduction 4 1.1 Heritability of cardiovascular diseases . .4 1.2 Genome wide era and complex diseases . .7 1.3 Functional genomics of disease associated loci . .8 1.4 Animal models for functional studies of CAD . 13 2 Genetic-Genomic Replication to Identify Candidate Mouse Atheroscle- rosis Modifier Genes 14 2.1 Introduction . 14 2.2 Methods . 15 2.2.1 Mouse and cell studies . 15 2.3 Results . 17 2.3.1 Atherosclerosis QTL replication in a new cross . 17 2.3.2 Protein coding differences between AKR and DBA mice resid- ing in ATH QTLs . 22 2.3.3 eQTL in bone marrow derived macrophages and endothelial cells 30 2.3.4 Macrophage eQTL replication between different crosses and dif- ferent platforms . 44 3 Transcriptome Analysis of Genes Regulated by Cholesterol Loading in Two Strains of Mouse Macrophages Associates Lysosome Pathway and ER Stress Response with Atherosclerosis Susceptibility 51 3.1 Introduction . -

(12) Patent Application Publication (10) Pub. No.: US 2011/0098188 A1 Niculescu Et Al
US 2011 0098188A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2011/0098188 A1 Niculescu et al. (43) Pub. Date: Apr. 28, 2011 (54) BLOOD BOMARKERS FOR PSYCHOSIS Related U.S. Application Data (60) Provisional application No. 60/917,784, filed on May (75) Inventors: Alexander B. Niculescu, Indianapolis, IN (US); Daniel R. 14, 2007. Salomon, San Diego, CA (US) Publication Classification (51) Int. Cl. (73) Assignees: THE SCRIPPS RESEARCH C40B 30/04 (2006.01) INSTITUTE, La Jolla, CA (US); CI2O I/68 (2006.01) INDIANA UNIVERSITY GOIN 33/53 (2006.01) RESEARCH AND C40B 40/04 (2006.01) TECHNOLOGY C40B 40/10 (2006.01) CORPORATION, Indianapolis, IN (52) U.S. Cl. .................. 506/9: 435/6: 435/7.92; 506/15; (US) 506/18 (57) ABSTRACT (21) Appl. No.: 12/599,763 A plurality of biomarkers determine the diagnosis of psycho (22) PCT Fled: May 13, 2008 sis based on the expression levels in a sample Such as blood. Subsets of biomarkers predict the diagnosis of delusion or (86) PCT NO.: PCT/US08/63539 hallucination. The biomarkers are identified using a conver gent functional genomics approach based on animal and S371 (c)(1), human data. Methods and compositions for clinical diagnosis (2), (4) Date: Dec. 22, 2010 of psychosis are provided. Human blood Human External Lines Animal Model External of Evidence changed in low vs. high Lines of Evidence psychosis (2pt.) Human postmortem s Animal model brai brain data (1 pt.) > Cite go data (1 p. Biomarker For Bonus 1 pt. Psychosis Human genetic 2 N linkage? association A all model blood data (1 pt.) data (1 p. -

Lipid Phosphate Phosphatase 3 in Vascular Pathophysiology
Atherosclerosis 271 (2018) 156e165 Contents lists available at ScienceDirect Atherosclerosis journal homepage: www.elsevier.com/locate/atherosclerosis Review article Lipid phosphate phosphatase 3 in vascular pathophysiology * Marco Busnelli a, , Stefano Manzini a, Cinzia Parolini a, Diana Escalante-Alcalde b, ** Giulia Chiesa a, a Department of Pharmacological and Biomolecular Sciences, Universita degli Studi di Milano, Milano, Italy b Instituto de Fisiología Celular, Division de Neurociencias Universidad Nacional Autonoma de Mexico, Cd. Mx., 04510, Mexico article info abstract Article history: LPP3 is an integral membrane protein belonging to a family of enzymes (LPPs) that display broad sub- Received 22 December 2017 strate specificity and catalyse dephosphorylation of several lipid substrates, including lysophosphatidic Received in revised form acid and sphingosine-1-phosphate. 2 February 2018 In mammals, the LPP family consists of three enzymes named LPP1, LPP2 and LPP3, which are encoded Accepted 20 February 2018 by three independent genes, PLPP1, PLPP2 and PLPP3, respectively (formerly known as PPAP2A, PPAP2C, Available online 2 March 2018 PPAP2B). These three enzymes, in vitro, do not seem to differ for catalytic activities and substrate pref- erences. However, in vivo targeted inactivation of the individual genes has indicated that the enzymes do Keywords: fi Lipid phosphate phosphatase 3 not have overlapping functions and that LPP3, speci cally, plays a crucial role in vascular development. fi Lysophosphatidic acid In 2011, two genome-wide association studies have identi ed PLPP3 as a novel locus associated with Sphingosine-1-phosphate coronary artery disease susceptibility. Shortly after these reports, tissue specific inactivation of PLPP3 in Genome-wide association studies mice highlighted a specific role for LPP3 in vascular pathophysiology and, more recently, in athero- Coronary artery disease sclerosis development.