DNA Metabarcoding of Stomach Contents
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prey Selection and Prey Handling in Two Raptors During the Breeding Season As Revealed by the Use of Video Monitoring
Norwegian University of Life Sciences Department of Ecology and Natural Resource Management (INA) Master Thesis 2015 30 credits Prey selection and prey handling in two raptors during the breeding season as revealed by the use of video monitoring Byttedyrvalg og byttedyrhåndtering hos to rovfugler i hekkeperioden, avslørt ved bruk av videoovervåking Ida Dihle A C K N O W L E D G E M E N T S First of all, I would like to thank Fylkesmannen i Oppland for the financial support. I would like to give a huge thanks to my brilliant supervisor Geir A. Sonerud who has spend hours identifying prey, helping with the statistical analyses and thoroughly reviewing my drafts. I am also very grateful to supervisor Ronny Steen, for all his effort regarding all stages of the fieldwork, his technical support throughout the process and providing helpful comments on my drafts. I would like to direct a big thanks to Vidar Selås, who contributed with his expertise when identifying prey. I am very grateful for the effort, inspiration and company during fieldwork given by Helge Grønnlien. The last weeks of recordings was made possible because of him. Dag S. Rusten also deserves a thanks, who along with Ronny Steen and Helge Grønnlien installed the cameras at the nests. My friends, Lene M. Rabben in particular, and family has been a big help throughout the process. I would like to thank my parents for providing equipment, and joining me in field, and Thea Dihle for punching numbers in the less inspiring periods of the writing process. -

Relating the 4-Year Lemming Population Cycle to a 3.8-Year Lunar Cycle and ENSO
Canadian Journal of Zoology Relating the 4-year lemming population cycle to a 3.8-year lunar cycle and ENSO Journal: Canadian Journal of Zoology Manuscript ID cjz-2018-0266.R3 Manuscript Type: Article Date Submitted by the 01-Jun-2019 Author: Complete List of Authors: Archibald, Herbert; Retired, ; None, None Is your manuscript invited for consideration in a Special Not applicable (regular submission) Issue?: Draft population cycle, lemming (Lemus spp. and Dicrostonyx spp.), zeitgeber, Keyword: lunar cycle, ENSO, Southern Oscillation Index (SOI) https://mc06.manuscriptcentral.com/cjz-pubs Page 1 of 37 Canadian Journal of Zoology Relating the 4-year lemming population cycle to a 3.8-year lunar cycle and ENSO Archibald, H.L. 632 Tee Hi Place, Medford, Wisconsin 54451 USA Correspondence: [email protected] Tel: 1-715-748-6536 Archibald, H. L. 2019. Relating the 4-year lemming population cycle to a 3.8-year lunar cycle and ENSO Draft 1 https://mc06.manuscriptcentral.com/cjz-pubs Canadian Journal of Zoology Page 2 of 37 Abstract Reported peak years of lemming (Lemmus spp. and Dicrostonyx spp.) and Arctic fox (Vulpes lagopus (Linnaeus, 1758)) abundance were compiled from the literature for 12 locations spanning 127 years. The mean period of the 34 reported lemming and Arctic fox cycles from 1868 to 1994 was 3.8 years, suggesting that the period of the 4-year cycle is actually 3.8 years. Peak population years were predicted using a simple model based on a 3.8-year lunar cycle. For nearly 130 years, reported years of peak abundance of lemmings and Arctic foxes were significantly correlated with and have persistently stayed in phase with predicted peak years of abundance. -

A-Nadachowski.Vp:Corelventura
Acta zoologica cracoviensia, 58(2): 181-194, Kraków, 31 December, 2015 Ó Institute of Systematics and Evolution of Animals, Pol. Acad. Sci., Kraków doi:10.3409/azc.58_2.181 NewLatePleistocenefaunalassemblagesfromPodhaleBasin, WesternCarpathians,Poland:preliminaryresults AdamNADACHOWSKI and Pawe³ VALDE-NOWAK Received: 05 October 2015. Accepted: 21 December 2015. Available online: 30 December 2015. NADACHOWSKI A., VALDE-NOWAK P. 2015. New Late Pleistocene faunal assem- blages from Podhale Basin, Western Carpathians, Poland: preliminary results. Acta zool. cracov., 58(2): 181-194. Abstract. Interdisciplinary studies in Ob³azowa Cave undertaken near its western entrance and in a new archaeological site in Cisowa Rock yielded finely layered sedimentary se- quences with an abundant fossil fauna of mainly small vertebrates and scarce archaeologi- cal finds. In this paper we present a preliminary palaeoenvironmental and palaeoclimatic reconstruction of the sequences dated around MIS 2/MIS 1 boundary on the basis of small mammal assemblages. The faunal changes appear to be gradual in spite of abrupt climatic changes expected to occur at that time. The environmental and faunal results are discussed with other sequences of similar age in Poland. Key words: fossil fauna, habitat and climate change, steppe-tundra, radiocarbon dating, Ob³azowa Cave. * Adam NADACHOWSKI, Institute of Systematics and Evolution of Animals Polish Acad- emy of Sciences, S³awkowska Str. 17, 31-016 Kraków, Poland. E-mail: [email protected] Pawe³ VALDE-NOWAK, Institute of Archaeology, Jagiellonian University, Go³êbia Str. 11, 31-007 Kraków, Poland. E-mail: [email protected] I.INTRODUCTION The Podhale Basin, also known as Orawa-Nowy Targ Basin, is located in the Western Carpathians, south Poland. -

Norway's Environmental Targets
Norway’s Environmental Targets Action for the environment 3 ————————— Priority areas and tools Useful for achieving our goals 4 websites: Priority areas www.miljo.no 01 Living seas and coastal environments 6 www.environment.no 02 Healthy lakes and rivers 10 www.klif.no 03 Rich and varied wetlands 14 www.dirnat.no 04 Forest diversity 18 www.npolar.no 05 Spectacular mountain landscapes 22 www.ra.no 06 A valuable cultural heritage 26 www.statkart.no 07 A good urban environment 30 08 The great outdoors 34 ————————— 09 An unpolluted environment 38 10 Clean air 42 11 A stable climate 46 Instruments Knowledge and geographical information 50 Legislation and planning procedures 52 International cooperation 54 Norway’s Environmental Targets was published in Norwegian in 2011. This translated version contains some updates and corrections, but has not been completely revised. 7U\NN$QGYRUG*UDILVN$6 Action for the environment Thirty years ago, acid rain was killing fish People have always left evidence of their in Norwegian rivers. Now, both salmon and activities behind them, altering the environ- trout are back again in many rivers thanks to ment for better or worse. Our cultural herit- concerted national and international efforts age does more than tell our history – it is also to cut emissions. part of a dynamic contemporary environ- ment. But it, too, is vulnerable: once build- Norway’s nature reaches from the deepest ings and cultural landscapes are destroyed seabed to the topmost mountain peaks, or removed, they are lost for ever. and from the thickest forest to the highest waterfall. -
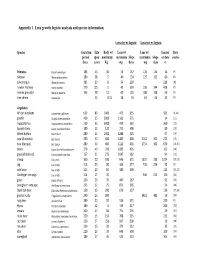
Appendix 1. Lens Growth Logistic Analysis and Species Information
Appendix 1. Lens growth logistic analysis and species information. Lens dry wt logistic Lens wet wt logistic Species Gestation Life Body wt Lens wt Lens wt Lenses Data period span maximum maximum Slope maximum Slope or data source days years Kg mg days mg days n Primates Papio hamadryas 183 45 30 55 167 178 124 16 PS baboon Macaca fascicularis 180 38 5 40 130 125 162 60 PS cynomolgus Alouatta caraya 185 20 8 34 220 228 [8] howler monkey Homo sapiens 270 115 5 45 359 198 244 608 PS human prenatal Macaca mulatta 165 38 12 60 125 188 108 50 PS tree shrew Tupaia glis 12 0.15 28 76 63 63 32 PS Ungulates African elephant Loxodonta a africana 659 80 5000 475 935 561 [9,10] giraffe Giraffa camelopardalis 460 35 2000 1163 571 34 [11] hipppotamus Hippopotamus amphibius 250 55 3000 410 362 360 [12] Spanish Ibex Capra pyrenaica Schinz 180 12 120 711 508 80 [13] Wood buffalo Bison bison 280 25 1000 1288 525 97 [14] cow (Australia) Bos taurus 280 30 600 1483 400 3162 302 231 [15] cow (Europe) Bos taurus 280 30 400 1122 401 2754 302 630 [16,17] zebra Equus burchelli antiquorum 370 40 350 1085 436 102 [18] gnu (wildebeest) Connochaetes taurinus 257 21 275 1047 362 94 [19] sheep Ovis aries 160 20 100 646 311 1622 239 1224 [20,21] pig Sus scrofa 115 25 80 364 277 736 254 92 PS wild boar Sus scrofa 115 20 84 389 309 113 [22,23] Gottingen minipigs Sus scrofa 114 17 35 646 211 100 [24] goat Capra hircus L 150 20 70 447 267 91 [25] pronghorn antelope Antilocapra americana 235 12 75 871 365 24 [26] black tail deer Odocoileus hemionus columbianus 150 15 200 -

Ice Age Animal Factsheets
Ice Age Animal Factsheets Scientists have found frozen woolly mammoths in Height: 3m (shoulder) cold places, like Russia. Weight: 6000 kg Some of these mammoths Diet: Herbivore were so complete that you Range: Eurasia & can see their hair and N. America muscles! WOOLLY MAMMOTH HERBIVORES: MEGAFAUNA The word “megafauna” means “big animal”. Many animals in the Ice Age were very large compared with creatures living today. WOOLLY RHINOCEROS Woolly rhinos were plant- eaters. They used their Height: 2m (shoulder) massive horns to scrape Weight: 2700 kg away ice and snow from Diet: Herbivore plants so they could feed Range: Eurasia (like a giant snow plough)! In Spain, there is a cave called Altamira which has Height: 2m (shoulder) beautiful Ice Age paintings Weight: 900 kg of bison. We also have Diet: Herbivore an Ice Age carv ing of a Range: Eurasia bison at Creswell Crags! STEPPE BISON HERBIVORES: COW-LIKE ANIMALS Aurochs and bison were large animals which looked a lot like bulls. Humans during the Ice Age would have hunted them for their meat and skins. Bison were one of the most popular The farm cows we have choicesAUROCHS of subject in Stone Age art. today are related to the Height: 1.8m (shoulder) aurochs. Scientists think Weight: 700 kg humans began to tame Diet: Herbivore aurochs around ten thousand Range: Eurasia & years ago. Africa The saiga antelope has a very unusual nose which Height: 0.7m (shoulder) helps them live in cold places. Breathing Weight: 40 kg through their nose lets Diet: Herbivore them warm up cold air! Range: Eurasia SAIGA ANTELOPE HERBIVORES: GRAZERS Grazers are animals which eat food on the ground, usually grasses or herby plants. -

Southern Norway
Southern Norway 05.07.-25.07.2018 Ingo Weiss, [email protected] During a mixed trip to southern Norway I could observe the following mammals. I do not have time at the moment for a more detailed report, nor for my pictures, but maybe this is interesting to someone. Most sites can be found in Tveit: A birdwatcher´s guide to Norway. Otherwise, feel free to ask. Family Scientific Name German Name English Name Date Site Arvicolinae Clethrionomys glareolus Rötelmaus Bank Vole 10.07.2018 Elverum SE 11.07.2018 Elverum SE 23.07.2018 Trillemarka - Grunntjennsetra Clethrionomys rufocanus Graurötelmaus Grey-sided Vole 07.07.2018 Hardangervidda - Tinnhølenveien 15.07.2018 Dovrefjell - Kongsvold - Nystugudalen Lemmus lemmus Berglemming Norway Lemming 06.07.2018 Flogdalen - Suleskard 06.07.2018 Håkallervatn 06.07.2018 Roskreppfjorden 07.07.2018 Hardangervidda - Tinnhølenveien 08.07.2018 Hardangervidda - Trondsbunuten Microtus agrestis Erdmaus Field Vole 06.07.2018 Håkallervatn 06.07.2018 Roskreppfjorden 08.07.2018 Hardangervidda - Tinnhølenveien 08.07.2018 Hardangervidda - Trondsbunuten Microtus oeconomus Nordische Wühlmaus Root Vole 07.07.2018 Hardangervidda - Tinnhølenveien 08.07.2018 Hardangervidda - Tinnhølenveien 15.07.2018 Dovrefjell - Kongsvold - Nystugudalen 15.07.2018 Fokstumyra 16.07.2018 Dovrefjell - Stropsljodalen Myopus schisticolor Waldlemming Wood Lemming 10.07.2018 Elverum SE Bovidae Ovibos moschatus Moschusochse Muskox 15.07.2018 Dovrefjell - Nystugudalen 16.07.2018 Dovrefjell - Stropsljodalen Canidae Vulpes vulpes Rotfuchs Red Fox -

Phylogenetic Diversity of Arctic Vertebrate Herbivores
Phylogenetic Diversity of Arctic Vertebrate Herbivores Ina Åsnes Skjelbred Master of Science Submission date: June 2017 Supervisor: James Speed, IBI Co-supervisor: Michael David Martin, IBI Norwegian University of Science and Technology Department of Biology ACKNOWLEDGEMENTS This master thesis was written at the Norwegian University of Science and Technology (NTNU) University Museum. First of all, I would like to express my full gratitude to my excellent team of supervisors, James David M. Speed and Michael David Martin. Thank you both for the motivating discussions, support throughout the process, and always being available to answer all my questions. Specifically, I would like to make a special thank you to Michael for gathering the sequence data and performing the phylogenetic analyses used in this study, and Andrew H. Thornhill assisting him in this process. And a special thanks to James for gathering the distribution data used in this study, all help with R, and giving me the possibility to present my project at both the Herbivory Network Conference and the Norwegian Ecological Society Conference along the way. Thank you for giving me the opportunity to work with such an interesting project - it has truly been a wonderful experience! In the end, I would like to thank all my friends and fellow LUR-students in Trondheim who has supported me through the time working with this project. I would especially like to thank Jonas for always standing by my side supporting and motivating me, and to Kristian for the good discussions and input along the way. Ina Åsnes Skjelbred June 2017, Trondheim i ii ABSTRACT Understanding spatial diversity patterns, and factors shaping these patterns, is crucial for conservation planning, and is particularly important in areas undergoing severe climatic change. -

Around the Perspektywiczna Cave NEWSLETTER #2 (1/2017)
around the Perspektywiczna Cave NEWSLETTER #2 (1/2017) Scientific magazine publishing short reports on speleology, geomorphology, geology, available from: www.ing.pan.pl/str_prac/Krajcarz_Mc/Krajcarz_Maciej.htm climatology, archaeology, paleontology, biology, ecology and natural and cultural Editor in chief: Maciej T. Krajcarz PhD, Institute of Geological Sciences, heritage of Perspektywiczna Cave and its vicinities Polish Academy of Sciences, Twarda St. 51/55, 00-818 Warszawa, Poland e-mail [email protected] published on: 30.06.2017 2016 excavaon area in Perspektywiczna Cave A huge sandstone nodule by Maciej T. Krajcarz found in the sediments of Perspektywiczna Cave Instute of Geological Sciences, Polish Academy of Sciences, email: [email protected] by Maciej T. Krajcarz During 2016 season (June 10 - August 20) excavaon works Instute of Geological Sciences, Polish Academy of Sciences, email: [email protected] concentrated in trench „V” only (area around the lower entrance Sandstone nodules have been excavated from the sedimentary fill of and the lower chamber of the cave; for localizaon of trenches see Perspektywiczna Cave during each excavaon season, and from the Newsleer #1). Excavated area covered square meters: D/3-6 and enre profile: from the topmost soil-like layer No. 1 to the sands of E/4-6 in the area in front of the cave, and A/8-10 and B-C/10 inside layer No. 11 lying on the limestone bedrock. Usually the size of the cave. The sediments at square meter B/7 parally collapsed nodules ranges from 1 to several cenmeters. In 2016 excavaon during winter, and were carefully collected, but the undisturbed season the unusually huge nodule was found in layer No. -

ECM2011-Abstract Book.Pdf
Tuesday 19 July 09:00 - 16:00 EXCURSION to Parc des Félins of Melun (charged to participants upon reservation) 14:00 - 18:00 Registration of delegates (Atrium, Jussieu Campus) Opening session 18:00 - 20:00 (Amphitheatre F1, Jussieu Campus) 18:00 - 18:15 Welcome - Christiane Denys & Stéphane Aulagnier - Opening of the congress 18:15 - 19:00 François Moutou - “About mammals and diseases” 19:00 - 19:30 Sabrina Renaud - “Evo-Devo and morphometrics: an insight into the evolutionary processes” 19:30 - 20:00 François Bonhomme - “Of mice and men: what’s new since the Neolithics ?” 20:30 - 23:00 WELCOME RECEPTION in the Grande Galerie de l’Evolution Wednesday 20 July AMPHI ASTIER AMPHI HERPIN AMPHI 45A AMPHI 45B AMPHI 55A AMPHI 55B 1 - Subterranean 21 - Population 22 - General 19 - Ungulates 09:00 - 10:30 20 - Macroevolution 4A - Shrews mammals ecology session genomics 10:30 - 11:15 Coffee break / Visit of exhibition area (Atrium) 1 - Subterranean 21 - Population 22 - General 19 - Ungulates 11:15 - 12:35 20 - Macroevolution 4A - Shrews mammals ecology session genomics 12:35 - 14:00 Lunch (Administrative University Restaurant) / Visit of exhibition area (Atrium) 1 - Subterranean 15 - Postglacial 21 - Population 22 - General 19 - Ungulates 14:00 - 15:30 4A - Shrews mammals recolonisation ecology session genomics 15:30 - 16:30 Poster session (Atrium) 16:30 - 17:00 Coffee break / Visit of exhibition area (Atrium) 1 - Subterranean 15 - Postglacial 21 - Population 22 - General 19 - Ungulates 17:00 - 18:00 4A - Shrews mammals recolonisation ecology session -

Arctic Biodiversity Assessment
528 Arctic Biodiversity Assessment Protostrongylus stilesi, a lung nematode typical in Dall’s sheep Ovis dalli from the Brooks Range and Alaska Range of the western North American Arctic, and in muskoxen Ovibos moschatus in the Brooks Range and Arctic Coastal Plain of Alaska and Yukon Territories, Canada. Shown is the tail end of an adult male with characteristic copulatory structures which are important in diagnosis of these miniscule parasites. Photo: E.P. Hoberg. 50 µm 529 Chapter 15 Parasites Lead Authors Eric P. Hoberg and Susan J. Kutz Contributing Authors Joseph A. Cook, Kirill Galaktionov, Voitto Haukisalmi, Heikki Henttonen, Sauli Laaksonen, Arseny Makarikov and David J. Marcogliese Contents Summary ..............................................................530 I’ve seen that in caribou. Just a couple of years ago, 15.1. Introduction .....................................................530 » every slice through it, you’d see about 50 little white 15.2. Parasites and their importance in the North ......................532 round things. We were wondering what that was, so we 15.3. Status and knowledge ...........................................533 checked it out, and it was a tapeworm. The whole body 15.4. Ecosystem components in the North .............................534 was completely filled with tapeworms. Yeah. It’s unbe- lievable how they could actually still move and run and 15.5. Terrestrial ecosystems ............................................536 15.5.1. Mammals ..................................................536 their whole body just completely filled with tapeworms. 15.5.1.1. Ungulates ..........................................536 Village elder, Sachs Harbour, Canada, as related to S.J. Kutz. 15.5.1.2. Rodents ...........................................538 15.5.2. Terrestrial birds .............................................539 15.6. Freshwater ecosystems ..........................................539 15.6.1. Fishes ......................................................540 He’s saying that when we go harvesting caribou, moose, 15.6.2. -

ARCTIC BIRDS INTERNATIONAL BREEDING CONDITIONS SURVEY BULLETIN No
CAFF Technical Report No.21 April 2010 ARCTIC BIRDS INTERNATIONAL BREEDING CONDITIONS SURVEY BULLETIN No. 11 Acknowledgements tat, conulluptat,Quis The Conservation of Arctic Flora and Fauna (CAFF) is a Working Group of the Arctic Council. CAFF Designated Agencies: • Environment Canada, Ottawa, Canada • Finnish Ministry of the Environment, Helsinki, Finland • The Ministry of Domestic A"airs, Nature and Environment, Greenland • Faroese Museum of Natural History, Tórshavn, Faroe Islands (Kingdom of Denmark) • Icelandic Institute of Natural History, Reykjavik, Iceland • Directorate for Nature Management, Trondheim, Norway • Russian Federation Ministry of Natural Resources, Moscow, Russia • Swedish Environmental Protection Agency, Stockholm, Sweden • United States Department of the Interior, Fish and Wildlife Service, Anchorage, Alaska CAFF Permanent Participant Organisations: • Aleut International Association (AIA) • Arctic Athabaskan Council (AAC) • Gwich’in Council International (GCI) • Inuit Circumpolar Conference - (ICC) Greenland, Alaska and Canada • Russian Indigenous Peoples of the North (RAIPON) • Saami Council This publication should be cited as: Soloviev, M.Y. and Tomkovich, P.S. (eds.). 2010. Arctic Birds, #11. CAFF Technical Report No. 21, CAFF International Secretariat, Akureyri, Iceland. Cover photo by Vasiliy Baranyuk. Incubating Common Eider on Wrangel Island in July 2008. For more information please contact: CAFF International Secretariat Borgir, Nordurslod 600 Akureyri, Iceland Phone: +354 462-3350 Fax: +354 462-3390 Email: