Decomposition Profile Data Analysis of Multiple Drug Effects Identifies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cambridgeshire and Peterborough Joint Prescribing Group MEDICINE REVIEW
Cambridgeshire and Peterborough Joint Prescribing Group MEDICINE REVIEW Name of Medicine / Trimipramine (Surmontil®) Class (generic and brand) Licensed indication(s) Treatment of depressive illness, especially where sleep disturbance, anxiety or agitation are presenting symptoms. Sleep disturbance is controlled within 24 hours and true antidepressant action follows within 7 to 10 days. Licensed dose(s) Adults: For depression 50-75 mg/day initially increasing to 150-300 mg/day in divided doses or one dose at night. The maintenance dose is 75-150 mg/day. Elderly: 10-25 mg three times a day initially. The initial dose should be increased with caution under close supervision. Half the normal maintenance dose may be sufficient to produce a satisfactory clinical response. Children: Not recommended. Purpose of Document To review information currently available on this class of medicines, give guidance on potential use and assign a prescribing classification http://www.cambsphn.nhs.uk/CJPG/CurrentDrugClassificationTable.aspx Annual cost (FP10) 10mg three times daily: £6,991 25mg three times daily: £7,819 150mg daily: £7,410 300mg daily: £14,820 Alternative Treatment Options within Class Tricyclic Annual Cost CPCCG Formulary Classification Antidepressant (FP10) Amitriptyline (75mg) Formulary £36 Lofepramine (140mg) Formulary £146 Imipramine (75mg) Non-formulary £37 Clomipramine (75mg) Non-formulary £63 Trimipramine (75mg). TBC £7,819 Nortriptyline (75mg) Not Recommended (pain) £276 Doxepin (150mg) TBC £6,006 Dosulepin (75mg) Not Recommended (NICE DO NOT DO) £19 Dosages are based on possible maintenance dose and are not equivalent between medications Recommendation It is recommended to Cambridgeshire and Peterborough CCG JPG members and through them to local NHS organisations that the arrangements for use of trimipramine are in line with restrictions agreed locally for drugs designated as NOT RECOMMENDED:. -

Cyproheptadine
PATIENT & CAREGIVER EDUCATION Cyproheptadine This information from Lexicomp® explains what you need to know about this medication, including what it’s used for, how to take it, its side effects, and when to call your healthcare provider. What is this drug used for? It is used to ease allergy signs. It is used to treat hives. It may be given to you for other reasons. Talk with the doctor. What do I need to tell my doctor BEFORE I take this drug? For all patients taking this drug: If you have an allergy to cyproheptadine or any other part of this drug. If you are allergic to this drug; any part of this drug; or any other drugs, foods, or substances. Tell your doctor about the allergy and what signs you had. If you have any of these health problems: Bowel block, enlarged prostate, glaucoma, trouble passing urine, or ulcers in your stomach or bowel. If you are taking certain drugs used for depression like Cyproheptadine 1/9 isocarboxazid, phenelzine, or tranylcypromine, or drugs used for Parkinson’s disease like selegiline or rasagiline. If you are taking any of these drugs: Linezolid or methylene blue. If you are 65 or older. If you are breast-feeding. Do not breast-feed while you take this drug. Children: If your child is a premature baby or is a newborn. Do not give this drug to a premature baby or a newborn. This is not a list of all drugs or health problems that interact with this drug. Tell your doctor and pharmacist about all of your drugs (prescription or OTC, natural products, vitamins) and health problems. -

Low-Dose Doxepin for Treatment of Pruritus in Patients on Hemodialysis
DIALYSIS Low-Dose Doxepin for Treatment of Pruritus in Patients on Hemodialysis Fatemeh Pour-Reza-Gholi,1 Alireza Nasrollahi,2 Ahmad Firouzan,1 Ensieh Nasli Esfahani,1 Farhat Farrokhi3 1Department of Nephrology, Introduction. Pruritus is one of the frequent discomforting Shaheed Labbafinejad complications in patients with end-stage renal disease. We Medical Center & Urology and prospectively evaluated the effectiveness of doxepin, an H1-receptor Nephrology Research Center, antagonist of histamine, in patients with pruritus resistant to Shaheed Beheshti Medical University, Tehran, Iran conventional treatment. 2Department of Nephrology, Materials and Methods. A randomized controlled trial with a Shohada-e-Tajrish Hospital, crossover design was performed on 24 patients in whom other Shaheed Beheshti Medical etiologic factors of pruritus had been ruled out. They were assigned University, Tehran, Iran into 2 groups and received either placebo or oral doxepin, 10 mg, 3Urology and Nephrology Research Center, Shaheed twice a day for 1 week. After a 1-week washout period, the 2 groups Beheshti Medical University, were treated conversely. Subjective outcome was determined by Tehran, Iran asking the patients described their pruritus as completely improved, relatively improved, or remained unchanged/worsened. Keywords. pruritus, doxepin, Results. Complete resolution of pruritus was reported in end-stage renal disease, 14 patients (58.3%) with doxepin and 2 (8.3%) with placebo dialysis (P < .001). Relative improvement was observed in 7 (29.2%) and 4 (16.7%), respectively. Overall, the improving effect of doxepin on Original Paper pruritus was seen in 87.5% of the patients. Twelve patients (50.0%) complained of drowsiness that alleviated in all cases after 2 days in average. -

Psychotropic Medications Judicial Reference Guide
PSYCHOTROPIC MEDICATIONS JUDICIAL REFERENCE GUIDE (Revised Edition 7/15/10) PSYCHOTROPIC MEDICATIONS JUDICIAL REFERENCE GUIDE FIRST EDITION THE STEERING COMMITTEE ON FAMILIES AND CHILDREN IN THE COURT Distributed by Florida Supreme Court 500 South Duval Street Tallahassee, FL 32399-1900 (850) 488-0125 INTRODUCTION One of the toughest challenges facing our dependency courts is the mental health of our children. “In July 2003, the Florida Statewide Advocacy Council published a Red Item Report finding 55% of foster children…in the state of Florida had been put on powerful mind altering psychotropic drugs.”1 In order to assist in this regard, the Psychotherapeutic Medication Subcommittee of the Steering Committee on Families and Children in the Court of the Supreme Court of Florida compiled this resource guide to help judges have a better understanding of psychotropic medications and their interaction with other drugs and with mental health disorders. Recently, the tragic case of Gabriel Myers in 2009 highlighted the fact that a number of child deaths were linked to the off label use of anti-psychotic medications. This is of special concern to Dependency Judges who are ultimately responsible for children in Florida’s Foster Care system. The researchers used publically available data from the internet, FDA manufactures’ published guidelines, publically available non-copyrighted articles and Dr. Brenda Thompson graciously prepared the Psychotropic Medication Chart. Special thanks to Dr. Brenda Thompson, the Honorable Herbert J. Baumann, the Honorable Ralph C. Stoddard, General Magistrate Tracy Ellis, Avron Bernstein, Selena Schoonover, Daniel Ringhoff, Jovasha Lang and to the Members of the Psychotherapeutic Medication Subcommittee. -

Medications That May Interfere with Skin Testing
MEDICATIONS THAT MAY INTERFERE WITH SKIN TESTING • Due to continued advances, not all medications may be listed at time of printing. • For your safety and accurate results, at each visit, please list all your current medications (including non-prescription and those prescribed elsewhere). • It is important to let us know if you are pregnant or could be pregnant. STOP THESE MEDICATIONS FIVE DAYS BEFORE SKIN TESTING: ORAL ANTIHISTAMINES: ANTIHISTAMINE NOSE SPRAYS: • Allegra (Fexofenadine) • Astelin, Astepro, Dymista (Azelastine) • Benadryl (Diphenhydramine) • Patanase (Olopatadine) • Claritin, Alavert (Loratadine) • Clarinex (Desloratadine) ANTIHISTAMINE EYE DROPS: • Xyzal (Levocetirizine) • Alaway, Claritin, Zaditor, Zyrtec (Ketotifen) • Zyrtec (Cetirizine) • Bepreve (Bepotastine) • Cyproheptadine • Elestat (Epinastine) · All over-the-counter medications for allergy, cough, cold, sleep, • Emadine (Emedastine) or nausea that include: • Lastacaft (Alcaftadine) oAcrivastine (ex. Semprex) • Livostin (Levocabastine) oAzatadine (ex. Optimine, Trinalin) • Naphcon-A, Opcon-A, Visine-A oBrompheniramine (ex. Dimetapp) (Pheniramine) oCarbinoxamine (ex. Palgic, Arbinoxa) • Optivar (Azelastine) oChlorpheniramine (ex. Actifed, Aller-chlor, Chlor- • Pataday, Patanol (Olopatadine) Trimeton, Tylenol Allergy) oDimenhydrinate (ex. Dramamine) HEARTBURN MEDICATIONS (H2 BLOCKERS): oDiphenhydramine (ex. Unisom, Sominex, • Axid (Nizatidine) Triaminic, many with “PM” in the title) • Pepcid, Tums Dual Action (Famotidine) oDoxylamine (ex. Nyquil, Unisom) • Tagament -

Male Anorgasmia: from “No” to “Go!”
Male Anorgasmia: From “No” to “Go!” Alexander W. Pastuszak, MD, PhD Assistant Professor Center for Reproductive Medicine Division of Male Reproductive Medicine and Surgery Scott Department of Urology Baylor College of Medicine Disclosures • Endo – speaker, consultant, advisor • Boston Scientific / AMS – consultant • Woven Health – founder, CMO Objectives • Understand what delayed ejaculation (DE) and anorgasmia are • Review the anatomy and physiology relevant to these conditions • Review what is known about the causes of DE and anorgasmia • Discuss management of DE and anorgasmia Definitions Delayed Ejaculation (DE) / Anorgasmia • The persistent or recurrent delay, difficulty, or absence of orgasm after sufficient sexual stimulation that causes personal distress Intravaginal Ejaculatory Latency Time (IELT) • Normal (median) à 5.4 minutes (0.55-44.1 minutes) • DE à mean IELT + 2 SD = 25 minutes • Incidence à 2-11% • Depends in part on definition used J Sex Med. 2005; 2: 492. Int J Impot Res. 2012; 24: 131. Ejaculation • Separate event from erection! • Thus, can occur in the ABSENCE of erection! Periurethral muscle Sensory input - glans (S2-4) contraction Emission Vas deferens contraction Sympathetic input (T12-L1) SV, prostate contraction Bladder neck contraction Expulsion Bulbocavernosus / Somatic input (S1-3) spongiosus contraction Projectile ejaculation J Sex Med. 2011; 8 (Suppl 4): 310. Neurochemistry Sexual Response Areas of the Brain • Pons • Nucleus paragigantocellularis Neurochemicals • Norepinephrine, serotonin: • Inhibit libido, -

Serotonin Syndrome How to Avoid, Identify, &
Serotonin syndrome How to avoid, identify, & As the list of serotonergic agents grows, recognizing hyperthermic states and potentially dangerous drug combinations is critical to our patients’ safety. 14 Current VOL. 2, NO. 5 / MAY 2003 p SYCHIATRY Current p SYCHIATRY treat dangerous drug interactions Harvey Sternbach, MD Clinical professor of psychiatry UCLA Neuropsychiatric Institute Los Angeles, CA romptly identifying serotonin syn- drome and acting decisively can keep side effects at the mild end of the spec- Ptrum. Symptoms of this potentially dangerous syndrome range from minimal in patients starting selective serotonin reuptake inhibitors (SSRIs) to fatal in those combining monoamine oxidase inhibitors (MAOIs) with serotonergic agents. This article presents the latest evidence on how to: • reduce the risk of serotonin syndrome • recognize its symptoms • and treat patients with mild to life- threatening symptoms. WHAT IS SEROTONIN SYNDROME? Serotonin syndrome is characterized by changes in autonomic, neuromotor, and cognitive-behav- ioral function (Table 1) triggered by increased serotonergic stimulation. It typically results from pharmacodynamic and/or pharmacokinetic in- teractions between drugs that increase serotonin activity.1,2 continued VOL. 2, NO. 5 / MAY 2003 15 Serotonin Table 1 activity or reduced ability to How to recognize serotonin syndrome secrete endothelium-derived nitric oxide may diminish the System Clinical signs and symptoms ability to metabolize serotonin.2 Autonomic Diaphoresis, hyperthermia, hypertension, tachycardia, pupillary dilatation, nausea, POTENTIALLY DANGEROUS diarrhea, shivering COMBINATIONS Neuromotor Hyperreflexia, myoclonus, restlessness, MAOIs. Serotonin syndrome tremor, incoordination, rigidity, clonus, has been reported as a result of teeth chattering, trismus, seizures interactions between MAOIs— Cognitive-behavioral Confusion, agitation, anxiety, hypomania, including selegiline and insomnia, hallucinations, headache reversible MAO-A inhibitors (RIMAs)—and various sero- tonergic compounds. -

CENTRAL NERVOUS SYSTEM DEPRESSANTS Opioid Pain Relievers Anxiolytics (Also Belong to Psychiatric Medication Category) • Codeine (In 222® Tablets, Tylenol® No
CENTRAL NERVOUS SYSTEM DEPRESSANTS Opioid Pain Relievers Anxiolytics (also belong to psychiatric medication category) • codeine (in 222® Tablets, Tylenol® No. 1/2/3/4, Fiorinal® C, Benzodiazepines Codeine Contin, etc.) • heroin • alprazolam (Xanax®) • hydrocodone (Hycodan®, etc.) • chlordiazepoxide (Librium®) • hydromorphone (Dilaudid®) • clonazepam (Rivotril®) • methadone • diazepam (Valium®) • morphine (MS Contin®, M-Eslon®, Kadian®, Statex®, etc.) • flurazepam (Dalmane®) • oxycodone (in Oxycocet®, Percocet®, Percodan®, OxyContin®, etc.) • lorazepam (Ativan®) • pentazocine (Talwin®) • nitrazepam (Mogadon®) • oxazepam ( Serax®) Alcohol • temazepam (Restoril®) Inhalants Barbiturates • gases (e.g. nitrous oxide, “laughing gas”, chloroform, halothane, • butalbital (in Fiorinal®) ether) • secobarbital (Seconal®) • volatile solvents (benzene, toluene, xylene, acetone, naptha and hexane) Buspirone (Buspar®) • nitrites (amyl nitrite, butyl nitrite and cyclohexyl nitrite – also known as “poppers”) Non-Benzodiazepine Hypnotics (also belong to psychiatric medication category) • chloral hydrate • zopiclone (Imovane®) Other • GHB (gamma-hydroxybutyrate) • Rohypnol (flunitrazepam) CENTRAL NERVOUS SYSTEM STIMULANTS Amphetamines Caffeine • dextroamphetamine (Dexadrine®) Methelynedioxyamphetamine (MDA) • methamphetamine (“Crystal meth”) (also has hallucinogenic actions) • methylphenidate (Biphentin®, Concerta®, Ritalin®) • mixed amphetamine salts (Adderall XR®) 3,4-Methelynedioxymethamphetamine (MDMA, Ecstasy) (also has hallucinogenic actions) Cocaine/Crack -
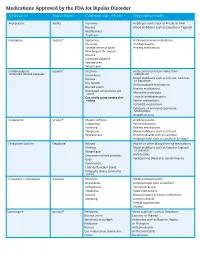
Medications Approved by the FDA for Bipolar Disorder Medication Brand Name Common Side Effects May Interact With
Medications Approved by the FDA for Bipolar Disorder Medication Brand Name Common side effects May interact with Aripiprazole Abilify® Insomnia Antidepressants such as Prozac or Paxil Nausea Mood stabilizers such as Equetro or Tegretol Restlessness Tiredness Asenapine Saphris® Sleepiness Antihypertensive medications Dizziness Antidepressants Strange sense of taste Anxiety medications Numbing of the mouth Nausea Increased appetite Feeling tired Weight gain Carbamazepine Equetro™ Dizziness Birth control pills (can make them extended release capsules Drowsiness ineffective) Mood stabilizers such as Lithium, Lamictal, Nausea or Depakote Dry mouth Anticonvulsant medications Blurred vision Anxiety medications Decreased white blood cell count Macrolide antibiotics Can rarely cause severe skin Tricyclic antidepressants rashes Cancer medications HIV/AIDS medications Cytotoxic or immunosuppressive Medications Grapefruit juice Cariprazine Vraylar® Muscle stiffness Antidepressants Indigestion Pain medications Vomiting Anxiety medications Sleepiness Mood stabilizers such as Lithium Restlessness Anticonvulsants such as Lamictal Antipsychotics such as Latuda or Seroquel Divalproex Sodium Depakote® Nausea Aspirin or other blood thinning medications Shaking Mood stabilizers such as Equetro,Tegretol, Weight gain or Lamictal Decrease in blood platelets Barbiturates Rash Cyclosporine (Neoral or Sandimmune) Pancreatitis Liver dysfunction (rare) Polycystic Ovary Syndrome (rare) Fluoxetine + Olanzapine Symbyax® Dizziness MAOI antidepressants Drowsiness Antipsychotics -

Trimipramine
PATIENT & CAREGIVER EDUCATION Trimipramine This information from Lexicomp® explains what you need to know about this medication, including what it’s used for, how to take it, its side effects, and when to call your healthcare provider. Brand Names: US Surmontil [DSC] Brand Names: Canada APO-Trimipramine Warning Drugs like this one have raised the chance of suicidal thoughts or actions in children and young adults. The risk may be greater in people who have had these thoughts or actions in the past. All people who take this drug need to be watched closely. Call the doctor right away if signs like low mood (depression), nervousness, restlessness, grouchiness, panic attacks, or changes in mood or actions are new or worse. Call the doctor right away if any thoughts or actions of suicide occur. This drug is not approved for use in children. Talk with the doctor. Trimipramine 1/10 What is this drug used for? It is used to treat low mood (depression). What do I need to tell my doctor BEFORE I take this drug? If you are allergic to this drug; any part of this drug; or any other drugs, foods, or substances. Tell your doctor about the allergy and what signs you had. If you have had a recent heart attack. If you have taken certain drugs for depression or Parkinson’s disease in the last 14 days. This includes isocarboxazid, phenelzine, tranylcypromine, selegiline, or rasagiline. Very high blood pressure may happen. If you are taking any of these drugs: Linezolid or methylene blue. This is not a list of all drugs or health problems that interact with this drug. -
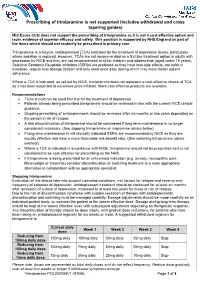
Prescribing of Trimipramine Is Not Supported (Includes Withdrawal and Cross Tapering Guides)
Prescribing of trimipramine is not supported (includes withdrawal and cross tapering guides) Mid Essex CCG does not support the prescribing of trimipramine as it is not a cost-effective option and lacks evidence of superior efficacy and safety. This position is supported by NHS England as part of the items which should not routinely be prescribed in primary care. Trimipramine is a tricyclic antidepressant (TCA) indicated for the treatment of depressive illness, particularly where sedation is required. However, TCAs are not recommended as a first line treatment option in adults with depression by NICE and they are not recommended at all for children and adolescents (aged under 18 years). Selective Serotonin Reuptake Inhibitors (SSRIs) are preferred as they have less side effects, are safer in overdose, require less dosage titration and only need once daily dosing which may mean better patient adherence. Where a TCA is indicated, as set out by NICE, trimipramine does not represent a cost-effective choice of TCA as it has been subjected to excessive price inflation. More cost effective products are available. Recommendations • TCAs should not be used first line for the treatment of depression. • Patients already being prescribed trimipramine should be reviewed in line with the current NICE clinical guidance. • Ongoing prescribing of antidepressants should be reviewed after six months or two years depending on the person’s risk of relapse. • A trial discontinuation of trimipramine should be considered if long-term maintenance is no longer considered necessary. (See stopping trimipramine or imipramine advice below) • If long-term maintenance is still clinically indicated SSRIs are recommended by NICE as they are equally effective and have a more favourable risk-benefit ratio. -

SELECTIVE SEROTONIN REUPTAKE INHIBITORS SEXUAL FUNCTION K. Demyttenaere0 and D. Vanderschueren" University Hospital Gasthui
* 1995 Elsevier Science B. V. All rights reserved. The Pharmacology of Sexual Function and Dysfunction J. Bancroft, editor 327 SELECTIVE SEROTONIN REUPTAKE INHIBITORS AND SEXUAL FUNCTION K. Demyttenaere0 and D. Vanderschueren" University Hospital Gasthuisberg (K.D. Leuven) ° Department of Psychiatry and Institute for Family and Sexological Sciences " Department of Endocrinology - Andrology Unit Herestraat 49 - B 3000 Leuven, Belgium dedicated to dr. Hugo De Cuyper (18.1.1946 - 13.9.1994) Introduction Selective Serotonin Reuptake Inhibitors (SSRIs : fluvoxamine -Floxyfral® or Fevarin®-, fluoxetine -Prozac®-, sertraline - Serlain® or Zoloft®-, paroxetine -Seroxat® or Paxil®- and citalopram -Cipramil®-) are potent and competitive inhibitors of the high affinity neuronal re-uptake mechanism for serotonin. These drugs also inhibit the re-uptake of noradrenaline and, generally to a lesser extent, dopamine, at higher concentrations. Paroxetine is the most potent serotonin uptake inhibitor in vitro of the drugs investigated, and citalopram is the most selective inhibitor of serotonin re-uptake, with paroxetine being more selective than fluoxetine, fluvoxamine and sertraline. Of the drugs investigated, only sertraline is a more potent inhibitor of dopamine compared with noradrenaline uptake. SSRIs are not only as effective as the tricyclic antidepressants in the treatment of major depression but are also effective in the treatment of obsessive-compulsive disorder, panic disorder, eating disorders and premenstrual syndrome (1). Although the latency of onset to therapeutic response seems to be somewhat longer, most psychiatrists consider SSRIs the initital drug of choice in the treatment of depressive disorders. Indeed, these drugs are usually better tolerated since they do not produce muscarinic, histaminic and alpha adrenergic side effects.