SUPPLEMENTARY INFORMATION a High Resolution
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

View of HER2: Human Epidermal Growth Factor Receptor 2; TNBC: Triple-Negative Breast Resistance to Systemic Therapy in Patients with Breast Cancer
Wen et al. Cancer Cell Int (2018) 18:128 https://doi.org/10.1186/s12935-018-0625-9 Cancer Cell International PRIMARY RESEARCH Open Access Sulbactam‑enhanced cytotoxicity of doxorubicin in breast cancer cells Shao‑hsuan Wen1†, Shey‑chiang Su2†, Bo‑huang Liou3, Cheng‑hao Lin1 and Kuan‑rong Lee1* Abstract Background: Multidrug resistance (MDR) is a major obstacle in breast cancer treatment. The predominant mecha‑ nism underlying MDR is an increase in the activity of adenosine triphosphate (ATP)-dependent drug efux trans‑ porters. Sulbactam, a β-lactamase inhibitor, is generally combined with β-lactam antibiotics for treating bacterial infections. However, sulbactam alone can be used to treat Acinetobacter baumannii infections because it inhibits the expression of ATP-binding cassette (ABC) transporter proteins. This is the frst study to report the efects of sulbactam on mammalian cells. Methods: We used the breast cancer cell lines as a model system to determine whether sulbactam afects cancer cells. The cell viabilities in the present of doxorubicin with or without sulbactam were measured by MTT assay. Protein identities and the changes in protein expression levels in the cells after sulbactam and doxorubicin treatment were determined using LC–MS/MS. Real-time reverse transcription polymerase chain reaction (real-time RT-PCR) was used to analyze the change in mRNA expression levels of ABC transporters after treatment of doxorubicin with or without sulbactam. The efux of doxorubicin was measures by the doxorubicin efux assay. Results: MTT assay revealed that sulbactam enhanced the cytotoxicity of doxorubicin in breast cancer cells. The results of proteomics showed that ABC transporter proteins and proteins associated with the process of transcription and initiation of translation were reduced. -

Crystal Structure of Human Gadd45 Reveals an Active Dimer
Protein Cell 2011, 2(10): 814–826 Protein & Cell DOI 10.1007/s13238-011-1090-6 RESEARCH ARTICLE Crystal structure of human Gadd45 reveals an active dimer 1,2,3* 1* 4 4 1 1 3 Wenzheng Zhang , Sheng Fu , Xuefeng Liu , Xuelian Zhao✉ , Wenchi Zhang✉ , Wei Peng , Congying Wu , Yuanyuan Li3, Xuemei Li1, Mark Bartlam1,2, Zong-Hao Zeng1 , Qimin Zhan4 , Zihe Rao1,2,3 1 National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China 2 Tianjin Key Laboratory of Protein Science, College of Life Sciences, Nankai University, Tianjin 300071, China 3 Laboratory of Structural Biology, Tsinghua University, Beijing 100084, China 4 State Key Laboratory of Molecular Oncology, Cancer Institute, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, China ✉ Correspondence: [email protected] (Z.-H. Zeng), [email protected] (Q. Zhan) Received August 23, 2011 Accepted September 19, 2011 ABSTRACT INTRODUCTION The human Gadd45 protein family plays critical roles in The human Gadd45 family of growth arrest and DNA DNA repair, negative growth control, genomic stability, damage-inducible proteins has three members, Gadd45α, cell cycle checkpoints and apoptosis. Here we report the -β, and -γ, grouped together on the basis of their amino acid crystal structure of human Gadd45, revealing a unique sequences and functional similarities (Vairapandi et al., 1996; dimer formed via a bundle of four parallel helices, Takekawa and Saito, 1998; Nakayama et al., 1999). The involving the most conserved residues among the genes encoding the α, β and γ isoforms of Gadd45 are Gadd45 isoforms. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -
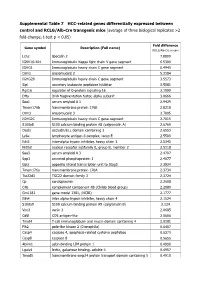
Supplemental Table 7 HCC-Related Genes Differentially Expressed Between Control and RCLG/Alb-Cre Transgenic Mice
Supplemental Table 7 HCC-related genes differentially expressed between control and RCLG/Alb-Cre transgenic mice (average of three biological replicates >2 fold-change, t-test p < 0.05) Fold difference Gene symbol Description (Full name) (RCLG/Alb-Cre vs con) Lcn2 lipocalin 2 7.8899 IGKV16-104 Immunoglobulin Kappa light chain V gene segment 6.5300 IGHG1 Immunoglobulin heavy chain C gene segment 6.4945 Orm2 orosomucoid 2 5.3184 IGHG2B Immunoglobulin heavy chain C gene segment 3.5573 Slpi secretory leukocyte peptidase inhibitor 3.5081 Rgs16 regulator of G-protein signaling 16 3.1999 Dffa DNA fragmentation factor, alpha subunit 3.0666 Saa1 serum amyloid A 1 2.9429 Tmem176b transmembrane protein 176B 2.8218 Orm3 orosomucoid 3 2.7805 IGHG2C Immunoglobulin heavy chain C gene segment 2.7015 S100a8 S100 calcium binding protein A8 (calgranulin A) 2.6769 Ocel1 occludin/ELL domain containing 1 2.6553 Ly6e lymphocyte antigen 6 complex, locus E 2.5509 Itih3 inter-alpha trypsin inhibitor, heavy chain 3 2.5345 Nr0b2 nuclear receptor subfamily 0, group B, member 2 2.5118 Saa3 serum amyloid A 3 2.4707 Spp1 secreted phosphoprotein 1 2.4077 Gats opposite strand transcription unit to Stag3 2.3934 Tmem176a transmembrane protein 176A 2.3734 Tsc22d3 TSC22 domain family 3 2.3724 Cp ceruloplasmin 2.2608 C4b complement component 4B (Childo blood group) 2.2089 Gm1381 gene model 1381, (NCBI) 2.1777 Itih4 inter alpha-trypsin inhibitor, heavy chain 4 2.1524 S100a9 S100 calcium binding protein A9 (calgranulin B) 2.124 Vnn3 vanin 3 2.0685 Cd5l CD5 antigen-like 2.0606 -

Open Dogan Phdthesis Final.Pdf
The Pennsylvania State University The Graduate School Eberly College of Science ELUCIDATING BIOLOGICAL FUNCTION OF GENOMIC DNA WITH ROBUST SIGNALS OF BIOCHEMICAL ACTIVITY: INTEGRATIVE GENOME-WIDE STUDIES OF ENHANCERS A Dissertation in Biochemistry, Microbiology and Molecular Biology by Nergiz Dogan © 2014 Nergiz Dogan Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy August 2014 ii The dissertation of Nergiz Dogan was reviewed and approved* by the following: Ross C. Hardison T. Ming Chu Professor of Biochemistry and Molecular Biology Dissertation Advisor Chair of Committee David S. Gilmour Professor of Molecular and Cell Biology Anton Nekrutenko Professor of Biochemistry and Molecular Biology Robert F. Paulson Professor of Veterinary and Biomedical Sciences Philip Reno Assistant Professor of Antropology Scott B. Selleck Professor and Head of the Department of Biochemistry and Molecular Biology *Signatures are on file in the Graduate School iii ABSTRACT Genome-wide measurements of epigenetic features such as histone modifications, occupancy by transcription factors and coactivators provide the opportunity to understand more globally how genes are regulated. While much effort is being put into integrating the marks from various combinations of features, the contribution of each feature to accuracy of enhancer prediction is not known. We began with predictions of 4,915 candidate erythroid enhancers based on genomic occupancy by TAL1, a key hematopoietic transcription factor that is strongly associated with gene induction in erythroid cells. Seventy of these DNA segments occupied by TAL1 (TAL1 OSs) were tested by transient transfections of cultured hematopoietic cells, and 56% of these were active as enhancers. Sixty-six TAL1 OSs were evaluated in transgenic mouse embryos, and 65% of these were active enhancers in various tissues. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Evolution of the DAN Gene Family in Vertebrates
bioRxiv preprint doi: https://doi.org/10.1101/794404; this version posted June 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. RESEARCH ARTICLE Evolution of the DAN gene family in vertebrates Juan C. Opazo1,2,3, Federico G. Hoffmann4,5, Kattina Zavala1, Scott V. Edwards6 1Instituto de Ciencias Ambientales y Evolutivas, Facultad de Ciencias, Universidad Austral de Chile, Valdivia, Chile. 2David Rockefeller Center for Latin American Studies, Harvard University, Cambridge, MA 02138, USA. 3Millennium Nucleus of Ion Channels-Associated Diseases (MiNICAD). 4 Department of Biochemistry, Molecular Biology, Entomology, and Plant Pathology, Mississippi State University, Mississippi State, 39762, USA. Cite as: Opazo JC, Hoffmann FG, 5 Zavala K, Edwards SV (2020) Institute for Genomics, Biocomputing, and Biotechnology, Mississippi State Evolution of the DAN gene family in University, Mississippi State, 39762, USA. vertebrates. bioRxiv, 794404, ver. 3 peer-reviewed and recommended by 6 PCI Evolutionary Biology. doi: Department of Organismic and Evolutionary Biology, Harvard University, 10.1101/794404 Cambridge, MA 02138, USA. This article has been peer-reviewed and recommended by Peer Community in Evolutionary Biology Posted: 29 June 2020 doi: 10.24072/pci.evolbiol.100104 ABSTRACT Recommender: Kateryna Makova The DAN gene family (DAN, Differential screening-selected gene Aberrant in Neuroblastoma) is a group of genes that is expressed during development and plays fundamental roles in limb bud formation and digitation, kidney formation and morphogenesis and left-right axis specification. -

Expression Profiling of KLF4
Expression Profiling of KLF4 AJCR0000006 Supplemental Data Figure S1. Snapshot of enriched gene sets identified by GSEA in Klf4-null MEFs. Figure S2. Snapshot of enriched gene sets identified by GSEA in wild type MEFs. 98 Am J Cancer Res 2011;1(1):85-97 Table S1: Functional Annotation Clustering of Genes Up-Regulated in Klf4 -Null MEFs ILLUMINA_ID Gene Symbol Gene Name (Description) P -value Fold-Change Cell Cycle 8.00E-03 ILMN_1217331 Mcm6 MINICHROMOSOME MAINTENANCE DEFICIENT 6 40.36 ILMN_2723931 E2f6 E2F TRANSCRIPTION FACTOR 6 26.8 ILMN_2724570 Mapk12 MITOGEN-ACTIVATED PROTEIN KINASE 12 22.19 ILMN_1218470 Cdk2 CYCLIN-DEPENDENT KINASE 2 9.32 ILMN_1234909 Tipin TIMELESS INTERACTING PROTEIN 5.3 ILMN_1212692 Mapk13 SAPK/ERK/KINASE 4 4.96 ILMN_2666690 Cul7 CULLIN 7 2.23 ILMN_2681776 Mapk6 MITOGEN ACTIVATED PROTEIN KINASE 4 2.11 ILMN_2652909 Ddit3 DNA-DAMAGE INDUCIBLE TRANSCRIPT 3 2.07 ILMN_2742152 Gadd45a GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE 45 ALPHA 1.92 ILMN_1212787 Pttg1 PITUITARY TUMOR-TRANSFORMING 1 1.8 ILMN_1216721 Cdk5 CYCLIN-DEPENDENT KINASE 5 1.78 ILMN_1227009 Gas2l1 GROWTH ARREST-SPECIFIC 2 LIKE 1 1.74 ILMN_2663009 Rassf5 RAS ASSOCIATION (RALGDS/AF-6) DOMAIN FAMILY 5 1.64 ILMN_1220454 Anapc13 ANAPHASE PROMOTING COMPLEX SUBUNIT 13 1.61 ILMN_1216213 Incenp INNER CENTROMERE PROTEIN 1.56 ILMN_1256301 Rcc2 REGULATOR OF CHROMOSOME CONDENSATION 2 1.53 Extracellular Matrix 5.80E-06 ILMN_2735184 Col18a1 PROCOLLAGEN, TYPE XVIII, ALPHA 1 51.5 ILMN_1223997 Crtap CARTILAGE ASSOCIATED PROTEIN 32.74 ILMN_2753809 Mmp3 MATRIX METALLOPEPTIDASE -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

MTH1 Favors Mesothelioma Progression and Mediates Paracrine Rescue of Bystander Endothelium from Oxidative Damage
MTH1 favors mesothelioma progression and mediates paracrine rescue of bystander endothelium from oxidative damage Sophia F. Magkouta, … , Vassilis G. Gorgoulis, Ioannis T. Kalomenidis JCI Insight. 2020;5(12):e134885. https://doi.org/10.1172/jci.insight.134885. Research Article Angiogenesis Oncology Oxidative stress and inadequate redox homeostasis is crucial for tumor initiation and progression. MTH1 (NUDT1) enzyme prevents incorporation of oxidized dNTPs by sanitizing the deoxynucleoside triphosphate (dNTP) pool and is therefore vital for the survival of tumor cells. MTH1 inhibition has been found to inhibit the growth of several experimental tumors, but its role in mesothelioma progression remained elusive. Moreover, although MTH1 is nonessential to normal cells, its role in survival of host cells in tumor milieu, especially tumor endothelium, is unclear. We validated a clinically relevant MTH1 inhibitor (Karonudib) in mesothelioma treatment using human xenografts and syngeneic murine models. We show that MTH1 inhibition impedes mesothelioma progression and that inherent tumoral MTH1 levels are associated with a tumor’s response. We also identified tumor endothelial cells as selective targets of Karonudib and propose a model of intercellular signaling among tumor cells and bystander tumor endothelium. We finally determined the major biological processes associated with elevated MTH1 gene expression in human mesotheliomas. Find the latest version: https://jci.me/134885/pdf RESEARCH ARTICLE MTH1 favors mesothelioma progression and mediates paracrine rescue of bystander endothelium from oxidative damage Sophia F. Magkouta,1 Apostolos G. Pappas,1 Photene C. Vaitsi,1 Panagiotis C. Agioutantis,1 Ioannis S. Pateras,2 Charalampos A. Moschos,1 Marianthi P. Iliopoulou,1 Chrysavgi N. Kosti,1 Heleni V. -

REVIEW ARTICLE the Genetics of Autism
REVIEW ARTICLE The Genetics of Autism Rebecca Muhle, BA*; Stephanie V. Trentacoste, BA*; and Isabelle Rapin, MD‡ ABSTRACT. Autism is a complex, behaviorally de- tribution of a few well characterized X-linked disorders, fined, static disorder of the immature brain that is of male-to-male transmission in a number of families rules great concern to the practicing pediatrician because of an out X-linkage as the prevailing mode of inheritance. The astonishing 556% reported increase in pediatric preva- recurrence rate in siblings of affected children is ϳ2% to lence between 1991 and 1997, to a prevalence higher than 8%, much higher than the prevalence rate in the general that of spina bifida, cancer, or Down syndrome. This population but much lower than in single-gene diseases. jump is probably attributable to heightened awareness Twin studies reported 60% concordance for classic au- and changing diagnostic criteria rather than to new en- tism in monozygotic (MZ) twins versus 0 in dizygotic vironmental influences. Autism is not a disease but a (DZ) twins, the higher MZ concordance attesting to ge- syndrome with multiple nongenetic and genetic causes. netic inheritance as the predominant causative agent. By autism (the autistic spectrum disorders [ASDs]), we Reevaluation for a broader autistic phenotype that in- mean the wide spectrum of developmental disorders cluded communication and social disorders increased characterized by impairments in 3 behavioral domains: 1) concordance remarkably from 60% to 92% in MZ twins social interaction; 2) language, communication, and and from 0% to 10% in DZ pairs. This suggests that imaginative play; and 3) range of interests and activities. -

(12) Patent Application Publication (10) Pub. No.: US 2016/0289762 A1 KOH Et Al
US 201602897.62A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2016/0289762 A1 KOH et al. (43) Pub. Date: Oct. 6, 2016 (54) METHODS FOR PROFILIING AND Publication Classification QUANTITATING CELL-FREE RNA (51) Int. Cl. (71) Applicant: The Board of Trustees of the Leland CI2O I/68 (2006.01) Stanford Junior University, Palo Alto, (52) U.S. Cl. CA (US) CPC ....... CI2O 1/6883 (2013.01); C12O 2600/112 (2013.01); C12O 2600/118 (2013.01); C12O (72) Inventors: Lian Chye Winston KOH, Stanford, 2600/158 (2013.01) CA (US); Stephen R. QUAKE, Stanford, CA (US); Hei-Mun Christina FAN, Fremont, CA (US); Wenying (57) ABSTRACT PAN, Stanford, CA (US) The invention generally relates to methods for assessing a (21) Appl. No.: 15/034,746 neurological disorder by characterizing circulating nucleic acids in a blood sample. According to certain embodiments, (22) PCT Filed: Nov. 6, 2014 methods for S. a Nial disorder include (86). PCT No.: PCT/US2O14/064355 obtaining RNA present in a blood sample of a patient Suspected of having a neurological disorder, determining a S 371 (c)(1), level of RNA present in the sample that is specific to brain (2) Date: May 5, 2016 tissue, comparing the sample level of RNA to a reference O O level of RNA specific to brain tissue, determining whether a Related U.S. Application Data difference exists between the sample level and the reference (60) Provisional application No. 61/900,927, filed on Nov. level, and indicating a neurological disorder if a difference 6, 2013.