Crystal Structure of Human Gadd45 Reveals an Active Dimer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

View of HER2: Human Epidermal Growth Factor Receptor 2; TNBC: Triple-Negative Breast Resistance to Systemic Therapy in Patients with Breast Cancer
Wen et al. Cancer Cell Int (2018) 18:128 https://doi.org/10.1186/s12935-018-0625-9 Cancer Cell International PRIMARY RESEARCH Open Access Sulbactam‑enhanced cytotoxicity of doxorubicin in breast cancer cells Shao‑hsuan Wen1†, Shey‑chiang Su2†, Bo‑huang Liou3, Cheng‑hao Lin1 and Kuan‑rong Lee1* Abstract Background: Multidrug resistance (MDR) is a major obstacle in breast cancer treatment. The predominant mecha‑ nism underlying MDR is an increase in the activity of adenosine triphosphate (ATP)-dependent drug efux trans‑ porters. Sulbactam, a β-lactamase inhibitor, is generally combined with β-lactam antibiotics for treating bacterial infections. However, sulbactam alone can be used to treat Acinetobacter baumannii infections because it inhibits the expression of ATP-binding cassette (ABC) transporter proteins. This is the frst study to report the efects of sulbactam on mammalian cells. Methods: We used the breast cancer cell lines as a model system to determine whether sulbactam afects cancer cells. The cell viabilities in the present of doxorubicin with or without sulbactam were measured by MTT assay. Protein identities and the changes in protein expression levels in the cells after sulbactam and doxorubicin treatment were determined using LC–MS/MS. Real-time reverse transcription polymerase chain reaction (real-time RT-PCR) was used to analyze the change in mRNA expression levels of ABC transporters after treatment of doxorubicin with or without sulbactam. The efux of doxorubicin was measures by the doxorubicin efux assay. Results: MTT assay revealed that sulbactam enhanced the cytotoxicity of doxorubicin in breast cancer cells. The results of proteomics showed that ABC transporter proteins and proteins associated with the process of transcription and initiation of translation were reduced. -

Table 2. Significant
Table 2. Significant (Q < 0.05 and |d | > 0.5) transcripts from the meta-analysis Gene Chr Mb Gene Name Affy ProbeSet cDNA_IDs d HAP/LAP d HAP/LAP d d IS Average d Ztest P values Q-value Symbol ID (study #5) 1 2 STS B2m 2 122 beta-2 microglobulin 1452428_a_at AI848245 1.75334941 4 3.2 4 3.2316485 1.07398E-09 5.69E-08 Man2b1 8 84.4 mannosidase 2, alpha B1 1416340_a_at H4049B01 3.75722111 3.87309653 2.1 1.6 2.84852656 5.32443E-07 1.58E-05 1110032A03Rik 9 50.9 RIKEN cDNA 1110032A03 gene 1417211_a_at H4035E05 4 1.66015788 4 1.7 2.82772795 2.94266E-05 0.000527 NA 9 48.5 --- 1456111_at 3.43701477 1.85785922 4 2 2.8237185 9.97969E-08 3.48E-06 Scn4b 9 45.3 Sodium channel, type IV, beta 1434008_at AI844796 3.79536664 1.63774235 3.3 2.3 2.75319499 1.48057E-08 6.21E-07 polypeptide Gadd45gip1 8 84.1 RIKEN cDNA 2310040G17 gene 1417619_at 4 3.38875643 1.4 2 2.69163229 8.84279E-06 0.0001904 BC056474 15 12.1 Mus musculus cDNA clone 1424117_at H3030A06 3.95752801 2.42838452 1.9 2.2 2.62132809 1.3344E-08 5.66E-07 MGC:67360 IMAGE:6823629, complete cds NA 4 153 guanine nucleotide binding protein, 1454696_at -3.46081884 -4 -1.3 -1.6 -2.6026947 8.58458E-05 0.0012617 beta 1 Gnb1 4 153 guanine nucleotide binding protein, 1417432_a_at H3094D02 -3.13334396 -4 -1.6 -1.7 -2.5946297 1.04542E-05 0.0002202 beta 1 Gadd45gip1 8 84.1 RAD23a homolog (S. -

Open Dogan Phdthesis Final.Pdf
The Pennsylvania State University The Graduate School Eberly College of Science ELUCIDATING BIOLOGICAL FUNCTION OF GENOMIC DNA WITH ROBUST SIGNALS OF BIOCHEMICAL ACTIVITY: INTEGRATIVE GENOME-WIDE STUDIES OF ENHANCERS A Dissertation in Biochemistry, Microbiology and Molecular Biology by Nergiz Dogan © 2014 Nergiz Dogan Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy August 2014 ii The dissertation of Nergiz Dogan was reviewed and approved* by the following: Ross C. Hardison T. Ming Chu Professor of Biochemistry and Molecular Biology Dissertation Advisor Chair of Committee David S. Gilmour Professor of Molecular and Cell Biology Anton Nekrutenko Professor of Biochemistry and Molecular Biology Robert F. Paulson Professor of Veterinary and Biomedical Sciences Philip Reno Assistant Professor of Antropology Scott B. Selleck Professor and Head of the Department of Biochemistry and Molecular Biology *Signatures are on file in the Graduate School iii ABSTRACT Genome-wide measurements of epigenetic features such as histone modifications, occupancy by transcription factors and coactivators provide the opportunity to understand more globally how genes are regulated. While much effort is being put into integrating the marks from various combinations of features, the contribution of each feature to accuracy of enhancer prediction is not known. We began with predictions of 4,915 candidate erythroid enhancers based on genomic occupancy by TAL1, a key hematopoietic transcription factor that is strongly associated with gene induction in erythroid cells. Seventy of these DNA segments occupied by TAL1 (TAL1 OSs) were tested by transient transfections of cultured hematopoietic cells, and 56% of these were active as enhancers. Sixty-six TAL1 OSs were evaluated in transgenic mouse embryos, and 65% of these were active enhancers in various tissues. -

Evolution of the DAN Gene Family in Vertebrates
bioRxiv preprint doi: https://doi.org/10.1101/794404; this version posted June 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. RESEARCH ARTICLE Evolution of the DAN gene family in vertebrates Juan C. Opazo1,2,3, Federico G. Hoffmann4,5, Kattina Zavala1, Scott V. Edwards6 1Instituto de Ciencias Ambientales y Evolutivas, Facultad de Ciencias, Universidad Austral de Chile, Valdivia, Chile. 2David Rockefeller Center for Latin American Studies, Harvard University, Cambridge, MA 02138, USA. 3Millennium Nucleus of Ion Channels-Associated Diseases (MiNICAD). 4 Department of Biochemistry, Molecular Biology, Entomology, and Plant Pathology, Mississippi State University, Mississippi State, 39762, USA. Cite as: Opazo JC, Hoffmann FG, 5 Zavala K, Edwards SV (2020) Institute for Genomics, Biocomputing, and Biotechnology, Mississippi State Evolution of the DAN gene family in University, Mississippi State, 39762, USA. vertebrates. bioRxiv, 794404, ver. 3 peer-reviewed and recommended by 6 PCI Evolutionary Biology. doi: Department of Organismic and Evolutionary Biology, Harvard University, 10.1101/794404 Cambridge, MA 02138, USA. This article has been peer-reviewed and recommended by Peer Community in Evolutionary Biology Posted: 29 June 2020 doi: 10.24072/pci.evolbiol.100104 ABSTRACT Recommender: Kateryna Makova The DAN gene family (DAN, Differential screening-selected gene Aberrant in Neuroblastoma) is a group of genes that is expressed during development and plays fundamental roles in limb bud formation and digitation, kidney formation and morphogenesis and left-right axis specification. -

Expression Profiling of KLF4
Expression Profiling of KLF4 AJCR0000006 Supplemental Data Figure S1. Snapshot of enriched gene sets identified by GSEA in Klf4-null MEFs. Figure S2. Snapshot of enriched gene sets identified by GSEA in wild type MEFs. 98 Am J Cancer Res 2011;1(1):85-97 Table S1: Functional Annotation Clustering of Genes Up-Regulated in Klf4 -Null MEFs ILLUMINA_ID Gene Symbol Gene Name (Description) P -value Fold-Change Cell Cycle 8.00E-03 ILMN_1217331 Mcm6 MINICHROMOSOME MAINTENANCE DEFICIENT 6 40.36 ILMN_2723931 E2f6 E2F TRANSCRIPTION FACTOR 6 26.8 ILMN_2724570 Mapk12 MITOGEN-ACTIVATED PROTEIN KINASE 12 22.19 ILMN_1218470 Cdk2 CYCLIN-DEPENDENT KINASE 2 9.32 ILMN_1234909 Tipin TIMELESS INTERACTING PROTEIN 5.3 ILMN_1212692 Mapk13 SAPK/ERK/KINASE 4 4.96 ILMN_2666690 Cul7 CULLIN 7 2.23 ILMN_2681776 Mapk6 MITOGEN ACTIVATED PROTEIN KINASE 4 2.11 ILMN_2652909 Ddit3 DNA-DAMAGE INDUCIBLE TRANSCRIPT 3 2.07 ILMN_2742152 Gadd45a GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE 45 ALPHA 1.92 ILMN_1212787 Pttg1 PITUITARY TUMOR-TRANSFORMING 1 1.8 ILMN_1216721 Cdk5 CYCLIN-DEPENDENT KINASE 5 1.78 ILMN_1227009 Gas2l1 GROWTH ARREST-SPECIFIC 2 LIKE 1 1.74 ILMN_2663009 Rassf5 RAS ASSOCIATION (RALGDS/AF-6) DOMAIN FAMILY 5 1.64 ILMN_1220454 Anapc13 ANAPHASE PROMOTING COMPLEX SUBUNIT 13 1.61 ILMN_1216213 Incenp INNER CENTROMERE PROTEIN 1.56 ILMN_1256301 Rcc2 REGULATOR OF CHROMOSOME CONDENSATION 2 1.53 Extracellular Matrix 5.80E-06 ILMN_2735184 Col18a1 PROCOLLAGEN, TYPE XVIII, ALPHA 1 51.5 ILMN_1223997 Crtap CARTILAGE ASSOCIATED PROTEIN 32.74 ILMN_2753809 Mmp3 MATRIX METALLOPEPTIDASE -

MTH1 Favors Mesothelioma Progression and Mediates Paracrine Rescue of Bystander Endothelium from Oxidative Damage
MTH1 favors mesothelioma progression and mediates paracrine rescue of bystander endothelium from oxidative damage Sophia F. Magkouta, … , Vassilis G. Gorgoulis, Ioannis T. Kalomenidis JCI Insight. 2020;5(12):e134885. https://doi.org/10.1172/jci.insight.134885. Research Article Angiogenesis Oncology Oxidative stress and inadequate redox homeostasis is crucial for tumor initiation and progression. MTH1 (NUDT1) enzyme prevents incorporation of oxidized dNTPs by sanitizing the deoxynucleoside triphosphate (dNTP) pool and is therefore vital for the survival of tumor cells. MTH1 inhibition has been found to inhibit the growth of several experimental tumors, but its role in mesothelioma progression remained elusive. Moreover, although MTH1 is nonessential to normal cells, its role in survival of host cells in tumor milieu, especially tumor endothelium, is unclear. We validated a clinically relevant MTH1 inhibitor (Karonudib) in mesothelioma treatment using human xenografts and syngeneic murine models. We show that MTH1 inhibition impedes mesothelioma progression and that inherent tumoral MTH1 levels are associated with a tumor’s response. We also identified tumor endothelial cells as selective targets of Karonudib and propose a model of intercellular signaling among tumor cells and bystander tumor endothelium. We finally determined the major biological processes associated with elevated MTH1 gene expression in human mesotheliomas. Find the latest version: https://jci.me/134885/pdf RESEARCH ARTICLE MTH1 favors mesothelioma progression and mediates paracrine rescue of bystander endothelium from oxidative damage Sophia F. Magkouta,1 Apostolos G. Pappas,1 Photene C. Vaitsi,1 Panagiotis C. Agioutantis,1 Ioannis S. Pateras,2 Charalampos A. Moschos,1 Marianthi P. Iliopoulou,1 Chrysavgi N. Kosti,1 Heleni V. -

Supplementary Materials
Supplementary materials Supplementary Table S1: MGNC compound library Ingredien Molecule Caco- Mol ID MW AlogP OB (%) BBB DL FASA- HL t Name Name 2 shengdi MOL012254 campesterol 400.8 7.63 37.58 1.34 0.98 0.7 0.21 20.2 shengdi MOL000519 coniferin 314.4 3.16 31.11 0.42 -0.2 0.3 0.27 74.6 beta- shengdi MOL000359 414.8 8.08 36.91 1.32 0.99 0.8 0.23 20.2 sitosterol pachymic shengdi MOL000289 528.9 6.54 33.63 0.1 -0.6 0.8 0 9.27 acid Poricoic acid shengdi MOL000291 484.7 5.64 30.52 -0.08 -0.9 0.8 0 8.67 B Chrysanthem shengdi MOL004492 585 8.24 38.72 0.51 -1 0.6 0.3 17.5 axanthin 20- shengdi MOL011455 Hexadecano 418.6 1.91 32.7 -0.24 -0.4 0.7 0.29 104 ylingenol huanglian MOL001454 berberine 336.4 3.45 36.86 1.24 0.57 0.8 0.19 6.57 huanglian MOL013352 Obacunone 454.6 2.68 43.29 0.01 -0.4 0.8 0.31 -13 huanglian MOL002894 berberrubine 322.4 3.2 35.74 1.07 0.17 0.7 0.24 6.46 huanglian MOL002897 epiberberine 336.4 3.45 43.09 1.17 0.4 0.8 0.19 6.1 huanglian MOL002903 (R)-Canadine 339.4 3.4 55.37 1.04 0.57 0.8 0.2 6.41 huanglian MOL002904 Berlambine 351.4 2.49 36.68 0.97 0.17 0.8 0.28 7.33 Corchorosid huanglian MOL002907 404.6 1.34 105 -0.91 -1.3 0.8 0.29 6.68 e A_qt Magnogrand huanglian MOL000622 266.4 1.18 63.71 0.02 -0.2 0.2 0.3 3.17 iolide huanglian MOL000762 Palmidin A 510.5 4.52 35.36 -0.38 -1.5 0.7 0.39 33.2 huanglian MOL000785 palmatine 352.4 3.65 64.6 1.33 0.37 0.7 0.13 2.25 huanglian MOL000098 quercetin 302.3 1.5 46.43 0.05 -0.8 0.3 0.38 14.4 huanglian MOL001458 coptisine 320.3 3.25 30.67 1.21 0.32 0.9 0.26 9.33 huanglian MOL002668 Worenine -

The Genetic Factors of Bilaterian Evolution Peter Heger1*, Wen Zheng1†, Anna Rottmann1, Kristen a Panfilio2,3, Thomas Wiehe1
RESEARCH ARTICLE The genetic factors of bilaterian evolution Peter Heger1*, Wen Zheng1†, Anna Rottmann1, Kristen A Panfilio2,3, Thomas Wiehe1 1Institute for Genetics, Cologne Biocenter, University of Cologne, Cologne, Germany; 2Institute for Zoology: Developmental Biology, Cologne Biocenter, University of Cologne, Cologne, Germany; 3School of Life Sciences, University of Warwick, Gibbet Hill Campus, Coventry, United Kingdom Abstract The Cambrian explosion was a unique animal radiation ~540 million years ago that produced the full range of body plans across bilaterians. The genetic mechanisms underlying these events are unknown, leaving a fundamental question in evolutionary biology unanswered. Using large-scale comparative genomics and advanced orthology evaluation techniques, we identified 157 bilaterian-specific genes. They include the entire Nodal pathway, a key regulator of mesoderm development and left-right axis specification; components for nervous system development, including a suite of G-protein-coupled receptors that control physiology and behaviour, the Robo- Slit midline repulsion system, and the neurotrophin signalling system; a high number of zinc finger transcription factors; and novel factors that previously escaped attention. Contradicting the current view, our study reveals that genes with bilaterian origin are robustly associated with key features in extant bilaterians, suggesting a causal relationship. *For correspondence: [email protected] Introduction The taxon Bilateria consists of multicellular animals -

Comparative Transcriptome Analysis of Muscular Dystrophy Models Largemyd, Dmdmdx&Sol
European Journal of Human Genetics (2016) 24, 1301–1309 & 2016 Macmillan Publishers Limited, part of Springer Nature. All rights reserved 1018-4813/16 www.nature.com/ejhg ARTICLE Comparative transcriptome analysis of muscular dystrophy models Largemyd, Dmdmdx/Largemyd and Dmdmdx: what makes them different? Camila F Almeida, Poliana CM Martins and Mariz Vainzof* Muscular dystrophies (MD) are a clinically and genetically heterogeneous group of Mendelian diseases. The underlying pathophysiology and phenotypic variability in each form are much more complex, suggesting the involvement of many other genes. Thus, here we studied the whole genome expression profile in muscles from three mice models for MD, at different time points: Dmdmdx (mutation in dystrophin gene), Largemyd − / − (mutation in Large)andDmdmdx/Largemyd − / − (both mutations). The identification of altered biological functions can contribute to understand diseases and to find prognostic biomarkers and points for therapeutic intervention. We identified a substantial number of differentially expressed genes (DEGs) in each model, reflecting diseases' complexity. The main biological process affected in the three strains was immune system, accounting for the majority of enriched functional categories, followed by degeneration/regeneration and extracellular matrix remodeling processes. The most notable differences were in 21-day-old Dmdmdx, with a high proportion of DEGs related to its regenerative capacity. A higher number of positive embryonic myosin heavy chain (eMyHC) fibers confirmed this. The new Dmdmdx/Largemyd − / − model did not show a highly different transcriptome from the parental lineages, with a profile closer to Largemyd − / − , but not bearing the same regenerative potential as Dmdmdx.Thisisthefirst report about transcriptome profile of a mouse model for congenital MD and Dmdmdx/Largemyd. -

The RNA-Protein Interactome of Differentiated Kidney Tubular Epithelial Cells
BASIC RESEARCH www.jasn.org The RNA-Protein Interactome of Differentiated Kidney Tubular Epithelial Cells Michael Ignarski,1 Constantin Rill,1 Rainer W.J. Kaiser,1 Madlen Kaldirim,1 René Neuhaus,1 Reza Esmaillie,1 Xinping Li,2 Corinna Klein,3 Katrin Bohl,1 Maike Petersen,1 Christian K. Frese,3 Martin Höhne ,1 Ilian Atanassov,2 Markus M. Rinschen,1 Katja Höpker,1 Bernhard Schermer,1,4,5 Thomas Benzing,1,4,5 Christoph Dieterich,6,7 Francesca Fabretti,1 and Roman-Ulrich Müller 1,4,5 1Department II of Internal Medicine and Center for Molecular Medicine Cologne, University of Cologne, Faculty of Medicine and University Hospital of Cologne, Cologne, Germany; 2Proteomics Core Facility, Max Planck Institute for Biology of Ageing, Cologne, Germany; 3Proteomics Facility, Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases, 4Nephrolab, Cologne Excellence Cluster on Cellular Stress Responses in Aging- associated Diseases, Faculty of Medicine and University Hospital Cologne, and 5Systems Biology of Ageing Cologne, University of Cologne, Cologne, Germany; 6Department of Internal Medicine III, Klaus Tschira Institute for Integrative Computational Cardiology, University Hospital Heidelberg, Heidelberg, Germany; and 7German Center for Cardiovascular Research (DZHK)–Partner site, Heidelberg/Mannheim, Germany ABSTRACT Background RNA-binding proteins (RBPs) are fundamental regulators of cellular biology that affect all steps in the generation and processing of RNA molecules. Recent evidence suggests that regulation of RBPs that modulate both RNA stability and translation may have a profound effect on the proteome. However, regulation of RBPs in clinically relevant experimental conditions has not been studied systematically. Methods We used RNA interactome capture, a method for the global identification of RBPs to characterize the global RNA‐binding proteome (RBPome) associated with polyA-tailed RNA species in murine ciliated epithelial cells of the inner medullary collecting duct. -

Genomic Analysis of Allele-Specific Expression in the Mouse Liver
bioRxiv preprint doi: https://doi.org/10.1101/024588; this version posted August 13, 2015. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 TITLE: Genomic analysis of allele-specific expression in the mouse liver Ashutosh K. Pandey* and Robert W. Williams* *Department of Genetics, Genomics and Informatics, University of Tennessee Health Science Center, Memphis, TN, 38103 bioRxiv preprint doi: https://doi.org/10.1101/024588; this version posted August 13, 2015. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 2 RUNNING TITLE: Allele-specific expression in liver KEYWORDS: BXD, DBA/2J, haplotype-aware alignment, purifying selection, cis eQTL CORRESPONDING AUTHOR: Ashutosh K. Pandey 855 Monroe Avenue, Suite 512 Memphis, TN, 38163 Phone: 901-448-1761 Email addresses: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/024588; this version posted August 13, 2015. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 3 ABSTRACT Genetic differences in gene expression contribute significantly to phenotypic diversity and differences in disease susceptibility. In fact, the great majority of causal variants highlighted by genome-wide association are in non-coding regions that modulate expression. In order to quantify the extent of allelic differences in expression, we analyzed liver transcriptomes of isogenic F1 hybrid mice. Allele-specific expression (ASE) effects are pervasive and are detected in over 50% of assayed genes. -
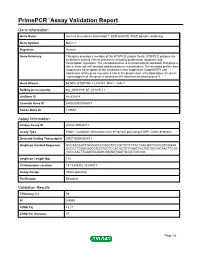
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name nucleus accumbens associated 1, BEN and BTB (POZ) domain containing Gene Symbol NACC1 Organism Human Gene Summary This gene encodes a member of the BTB/POZ protein family. BTB/POZ proteins are involved in several cellular processes including proliferation apoptosis and transcription regulation. The encoded protein is a transcriptional repressor that plays a role in stem cell self-renewal and pluripotency maintenance. The encoded protein also suppresses transcription of the candidate tumor suppressor Gadd45GIP1 and expression of this gene may play a role in the progression of multiple types of cancer. A pseudogene of this gene is located on the short arm of chromosome 9. Gene Aliases BEND8, BTBD14B, FLJ37383, NAC-1, NAC1 RefSeq Accession No. NC_000019.9, NT_011295.11 UniGene ID Hs.531614 Ensembl Gene ID ENSG00000160877 Entrez Gene ID 112939 Assay Information Unique Assay ID qHsaCIP0028717 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000292431 Amplicon Context Sequence GCCAACAGCTGCGGCACCGGCATCCGCTCTTCTACCAACGATCCCCGTCGGAA GCCCCTGGACAGCCGCGTGCTCCACGCTGTCAAGTACTACTGCCAGAACTTCGC CCCCAACTTCAAGGAGAGCGAGATGAATGCCATCGCGG Amplicon Length (bp) 115 Chromosome Location 19:13248302-13249017 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 98 R2 0.9995 cDNA Cq 19.17 cDNA Tm (Celsius) 87 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 27.88 Specificity (%) 100 Information