A TAF4 Coactivator Function for E Proteins That Involves Enhanced TFIID Binding
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Interaction and Functional Cooperation of the Cancer-Amplified Transcriptional Coactivator Activating Signal Cointegrator-2 and E2F-1 in Cell Proliferation
948 Vol. 1, 948–958, November 2003 Molecular Cancer Research Interaction and Functional Cooperation of the Cancer-Amplified Transcriptional Coactivator Activating Signal Cointegrator-2 and E2F-1 in Cell Proliferation Hee Jeong Kong,1 Hyun Jung Yu,2 SunHwa Hong,2 Min Jung Park,2 Young Hyun Choi,3 Won Gun An,4 Jae Woon Lee,5 and JaeHun Cheong2 1Laboratory of Molecular Growth Regulation, National Institute of Health, Bethesda, MD; 2Department of Molecular Biology, Pusan National University, Busan, Republic of Korea; 3Department of Biochemistry, College of Oriental Medicine, Dong-Eui University, Busan, Republic of Korea; 4Department of Biology, Kyungpook National University, DaeGu, Republic of Korea; and 5Department of Medicine/Endocrinology, Baylor College of Medicine Houston, TX. Abstract structure and recruitment of a transcription initiation complex Activating signal cointegrator-2 (ASC-2), a novel coac- containing RNA polymerase II to the promoter (1). Recent tivator, is amplified in several cancer cells and known to studies have shown that DNA-bound transcriptional activator interact with mitogenic transcription factors, including proteins accomplish these two tasks with the assistance of a serum response factor, activating protein-1, and nuclear class of proteins called transcriptional coactivators (2, 3). They factor-KB, suggesting the physiological role of ASC-2 in may be thought of as adaptors or components in a signaling the promotion of cell proliferation. Here, we show that pathway that transmits transcriptional activation signals from the expression pattern of ASC-2 was correlated with that DNA-bound activator proteins to the chromatin and transcrip- of E2F-1 for protein increases at G1 and S phase. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia. -

Original Article EP300 Regulates the Expression of Human Survivin Gene in Esophageal Squamous Cell Carcinoma
Int J Clin Exp Med 2016;9(6):10452-10460 www.ijcem.com /ISSN:1940-5901/IJCEM0023383 Original Article EP300 regulates the expression of human survivin gene in esophageal squamous cell carcinoma Xiaoya Yang, Zhu Li, Yintu Ma, Xuhua Yang, Jun Gao, Surui Liu, Gengyin Wang Department of Blood Transfusion, The Bethune International Peace Hospital of China PLA, Shijiazhuang 050082, Hebei, P. R. China Received January 6, 2016; Accepted March 21, 2016; Epub June 15, 2016; Published June 30, 2016 Abstract: Survivin is selectively up-regulated in various cancers including esophageal squamous cell carcinoma (ESCC). The underlying mechanism of survivin overexpression in cancers is needed to be further studied. In this study, we investigated the effect of EP300, a well known transcriptional coactivator, on survivin gene expression in human esophageal squamous cancer cell lines. We found that overexpression of EP300 was associated with strong repression of survivin expression at the mRNA and protein levels. Knockdown of EP300 increased the survivin ex- pression as indicated by western blotting and RT-PCR analysis. Furthermore, our results indicated that transcription- al repression mediated by EP300 regulates survivin expression levels via regulating the survivin promoter activity. Chromatin immunoprecipitation (ChIP) analysis revealed that EP300 was associated with survivin gene promoter. When EP300 was added to esophageal squamous cancer cells, increased EP300 association was observed at the survivin promoter. But the acetylation level of histone H3 at survivin promoter didn’t change after RNAi-depletion of endogenous EP300 or after overexpression of EP300. These findings establish a negative regulatory role for EP300 in survivin expression. Keywords: Survivin, EP300, transcription regulation, ESCC Introduction transcription factors and the basal transcrip- tion machinery, or by providing a scaffold for Survivin belongs to the inhibitor of apoptosis integrating a variety of different proteins [6]. -

Crystal Structure of the Bacteriophage T4 Late-Transcription Coactivator Gp33 with the Β-Subunit Flap Domain of Escherichia Coli RNA Polymerase
Crystal structure of the bacteriophage T4 late-transcription coactivator gp33 with the β-subunit flap domain of Escherichia coli RNA polymerase The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Twist, K.-A. F., E. A. Campbell, P. Deighan, S. Nechaev, V. Jain, E. P. Geiduschek, A. Hochschild, and S. A. Darst. 2011. “Crystal Structure of the Bacteriophage T4 Late-Transcription Coactivator gp33 with the -Subunit Flap Domain of Escherichia Coli RNA Polymerase.” Proceedings of the National Academy of Sciences 108 (50): 19961– 66. https://doi.org/10.1073/pnas.1113328108. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:41483152 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Crystal structure of the bacteriophage T4 late- transcription coactivator gp33 with the β-subunit flap domain of Escherichia coli RNA polymerase Kelly-Anne F. Twista,1, Elizabeth A. Campbella, Padraig Deighanb, Sergei Nechaevc,2, Vikas Jainc,3, E. Peter Geiduschekc, Ann Hochschildb, and Seth A. Darsta,4 aLaboratory of Molecular Biophysics, The Rockefeller University, 1230 York Avenue, New York, NY 10065; bDepartment of Microbiology and Immunobiology, Harvard Medical School, Boston, MA 02115; and cDivision of Biological Sciences, Section of Molecular Biology, University -

A Thyroid Hormone Receptor Coactivator Negatively Regulated by the Retinoblastoma Protein
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 9040–9045, August 1997 Biochemistry A thyroid hormone receptor coactivator negatively regulated by the retinoblastoma protein KAI-HSUAN CHANG*†,YUMAY CHEN*†,TUNG-TI CHEN*†,WEN-HAI CHOU*†,PHANG-LANG CHEN*, YEN-YING MA‡,TERESA L. YANG-FENG‡,XIAOHUA LENG§,MING-JER TSAI§,BERT W. O’MALLEY§, AND WEN-HWA LEE*¶ *Department of Molecular Medicine and Institute of Biotechnology, University of Texas Health Science Center at San Antonio, 15355 Lambda Drive, San Antonio, TX 78245; ‡Department of Genetics, Obstetrics, and Gynecology, Yale University School of Medicine, New Haven, CT 06510; and §Department of Cell Biology, Baylor College of Medicine, Houston, TX 77030 Contributed by Bert W. O’Malley, June 9, 1997 ABSTRACT The retinoblastoma protein (Rb) plays a E2F-1, a transcription factor important for the expression of critical role in cell proliferation, differentiation, and devel- several genes involved in cell cycle progression from G1 to S opment. To decipher the mechanism of Rb function at the (18). Rb inhibits E2F-1 activity by blocking its transactivation molecular level, we have systematically characterized a num- region (19–21). In contrast, Rb has been shown to have the ber of Rb-interacting proteins, among which is the clone C5 ability to increase the transactivating activity of the members described here, which encodes a protein of 1,978 amino acids of the CCAATyenhancer binding protein (CyEBP) family, and with an estimated molecular mass of 230 kDa. The corre- to be required for CyEBPs-dependent adipocyte and mono- sponding gene was assigned to chromosome 14q31, the same cytes differentiation (16–17). -

TAF10 Complex Provides Evidence for Nuclear Holo&Ndash;TFIID Assembly from Preform
ARTICLE Received 13 Aug 2014 | Accepted 2 Dec 2014 | Published 14 Jan 2015 DOI: 10.1038/ncomms7011 OPEN Cytoplasmic TAF2–TAF8–TAF10 complex provides evidence for nuclear holo–TFIID assembly from preformed submodules Simon Trowitzsch1,2, Cristina Viola1,2, Elisabeth Scheer3, Sascha Conic3, Virginie Chavant4, Marjorie Fournier3, Gabor Papai5, Ima-Obong Ebong6, Christiane Schaffitzel1,2, Juan Zou7, Matthias Haffke1,2, Juri Rappsilber7,8, Carol V. Robinson6, Patrick Schultz5, Laszlo Tora3 & Imre Berger1,2,9 General transcription factor TFIID is a cornerstone of RNA polymerase II transcription initiation in eukaryotic cells. How human TFIID—a megadalton-sized multiprotein complex composed of the TATA-binding protein (TBP) and 13 TBP-associated factors (TAFs)— assembles into a functional transcription factor is poorly understood. Here we describe a heterotrimeric TFIID subcomplex consisting of the TAF2, TAF8 and TAF10 proteins, which assembles in the cytoplasm. Using native mass spectrometry, we define the interactions between the TAFs and uncover a central role for TAF8 in nucleating the complex. X-ray crystallography reveals a non-canonical arrangement of the TAF8–TAF10 histone fold domains. TAF2 binds to multiple motifs within the TAF8 C-terminal region, and these interactions dictate TAF2 incorporation into a core–TFIID complex that exists in the nucleus. Our results provide evidence for a stepwise assembly pathway of nuclear holo–TFIID, regulated by nuclear import of preformed cytoplasmic submodules. 1 European Molecular Biology Laboratory, Grenoble Outstation, 6 rue Jules Horowitz, 38042 Grenoble, France. 2 Unit for Virus Host-Cell Interactions, University Grenoble Alpes-EMBL-CNRS, 6 rue Jules Horowitz, 38042 Grenoble, France. 3 Cellular Signaling and Nuclear Dynamics Program, Institut de Ge´ne´tique et de Biologie Mole´culaire et Cellulaire, UMR 7104, INSERM U964, 1 rue Laurent Fries, 67404 Illkirch, France. -

Structure and Mechanism of the RNA Polymerase II Transcription Machinery
Downloaded from genesdev.cshlp.org on October 9, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW Structure and mechanism of the RNA polymerase II transcription machinery Allison C. Schier and Dylan J. Taatjes Department of Biochemistry, University of Colorado, Boulder, Colorado 80303, USA RNA polymerase II (Pol II) transcribes all protein-coding ingly high resolution, which has rapidly advanced under- genes and many noncoding RNAs in eukaryotic genomes. standing of the molecular basis of Pol II transcription. Although Pol II is a complex, 12-subunit enzyme, it lacks Structural biology continues to transform our under- the ability to initiate transcription and cannot consistent- standing of complex biological processes because it allows ly transcribe through long DNA sequences. To execute visualization of proteins and protein complexes at or near these essential functions, an array of proteins and protein atomic-level resolution. Combined with mutagenesis and complexes interact with Pol II to regulate its activity. In functional assays, structural data can at once establish this review, we detail the structure and mechanism of how enzymes function, justify genetic links to human dis- over a dozen factors that govern Pol II initiation (e.g., ease, and drive drug discovery. In the past few decades, TFIID, TFIIH, and Mediator), pausing, and elongation workhorse techniques such as NMR and X-ray crystallog- (e.g., DSIF, NELF, PAF, and P-TEFb). The structural basis raphy have been complemented by cryoEM, cross-linking for Pol II transcription regulation has advanced rapidly mass spectrometry (CXMS), and other methods. Recent in the past decade, largely due to technological innova- improvements in data collection and imaging technolo- tions in cryoelectron microscopy. -

EZH2 Reduction Is an Essential Mechanoresponse for The
Li et al. Cell Death and Disease (2020) 11:757 https://doi.org/10.1038/s41419-020-02963-3 Cell Death & Disease ARTICLE Open Access EZH2 reduction is an essential mechanoresponse for the maintenance of super-enhancer polarization against compressive stress in human periodontal ligament stem cells Qian Li1,XiwenSun2,YunyiTang2, Yanan Qu2, Yanheng Zhou1 and Yu Zhang2 Abstract Despite the ubiquitous mechanical cues at both spatial and temporal dimensions, cell identities and functions are largely immune to the everchanging mechanical stimuli. To understand the molecular basis of this epigenetic stability, we interrogated compressive force-elicited transcriptomic changes in mesenchymal stem cells purified from human periodontal ligament (PDLSCs), and identified H3K27me3 and E2F signatures populated within upregulated and weakly downregulated genes, respectively. Consistently, expressions of several E2F family transcription factors and EZH2, as core methyltransferase for H3K27me3, decreased in response to mechanical stress, which were attributed to force-induced redistribution of RB from nucleoplasm to lamina. Importantly, although epigenomic analysis on H3K27me3 landscape only demonstrated correlating changes at one group of mechanoresponsive genes, we observed a genome-wide destabilization of super-enhancers along with aberrant EZH2 retention. These super- enhancers were tightly bounded by H3K27me3 domain on one side and exhibited attenuating H3K27ac deposition fl 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; and attening H3K27ac peaks along with compensated EZH2 expression after force exposure, analogous to increased H3K27ac entropy or decreased H3K27ac polarization. Interference of force-induced EZH2 reduction could drive actin filaments dependent spatial overlap between EZH2 and super-enhancers and functionally compromise the multipotency of PDLSC following mechanical stress. These findings together unveil a specific contribution of EZH2 reduction for the maintenance of super-enhancer stability and cell identity in mechanoresponse. -

Supplementary Tables Supplementary Table S1. Treatment Protocols
Supplementary Tables Supplementary Table S1. Treatment protocols pediatric AML patients. Number of Treatment patients ADE-based regimen 64 ADE with bortezomib 14 ADE with mylotarg 6 APL-like treatment 2 TMD-like treatment 1 Relapse therapy 2 Misc treatment/novel agents 5 Unknown 2 Supplemental Table S2: Patient characteristics for the 73 pediatric acute lymphoblastic leukemia patients Characteristics N, % Number of cases 73 ALL subtype Pre-B ALL 57 (78) T-ALL 16 (22) Age, y, median (range) 7.3 (0.2-18.0) Gender Male 40 (55) Female 33 (45) Declared Ethnicity Caucasian 61 (84) Hispanic 44 (72) Non-Hispanic 17 (28) Black American 6 (8) Asian 4 (5) Mixed 2 (3) Hispanic 1 (1) ‘‘SNP’’ Ethnicity European 9 (12) African 6 (8) American Indian 38 (52) Asian 1 (1) Not done 19 (26) Cytogenetics Favorable 15 (21) Intermediate 42 (58) Unfavorable 15 (21) Unknown 1 (1) Risk Group Low Risk 4 (5) Standard/ Intermediate Risk 29 (40) High/Very High Risk 40 (55) CNS status CNS-1 46 (63) CNS-2 20 (27) CNS-3 6 (8) Unknown 2 (3) Response Complete remission 67 (92) Resistant 4 (5) Fail 2 (3) Alive 63 (86) Supplementary Table S3. The table shows the ‘‘Rosetta Stone’’ of the antibody and protein nomenclature, and the R2 for the antibody validation and the primary and secondary antibody dilutions. Secondary, all antibodies that were used in combination with information about the manufacturer of each antibody, the antibody source, and catalog number are listed. Functional Protein Name Rosetta Stone RPPA Staining Details Category Functional Effect Common Name RPPA Antibody Huge Name (added MiMI Antibody R2, WB vs. -

Rapid Isolation and Identification of Bacteriophage T4-Encoded Modifications of Escherichia Coli RNA Polymerase
Rapid Isolation and Identification of Bacteriophage T4-Encoded Modifications of Escherichia coli RNA Polymerase: A Generic Method to Study Bacteriophage/Host Interactions Lars F. Westblade,† Leonid Minakhin,‡ Konstantin Kuznedelov,‡ Alan J. Tackett,†,§ Emmanuel J. Chang,†,4 Rachel A. Mooney,⊥ Irina Vvedenskaya,‡ Qing Jun Wang,† David Fenyö,† Michael P. Rout,† Robert Landick,⊥ Brian T. Chait,† Konstantin Severinov,‡ and Seth A. Darst*,† The Rockefeller University, New York, New York 10065, Waksman Institute for Microbiology and Department of Molecular Biology and Biochemistry, Rutgers, The State University of New Jersey, Piscataway, New Jersey 08854, Department of Biochemistry and Molecular Biology, University of Arkansas for Medical Sciences, Little Rock, Arkansas 72205, Department of Natural Sciences, York College/City University of New York, Jamaica, New York 11451, and Departments of Biochemistry and Bacteriology, University of Wisconsin-Madison, Madison, Wisconsin 53706 Received July 19, 2007 Bacteriophages are bacterial viruses that infect bacterial cells, and they have developed ingenious mechanisms to modify the bacterial RNA polymerase. Using a rapid, specific, single-step affinity isolation procedure to purify Escherichia coli RNA polymerase from bacteriophage T4-infected cells, we have identified bacteriophage T4-dependent modifications of the host RNA polymerase. We suggest that this methodology is broadly applicable for the identification of bacteriophage-dependent alterations of the host synthesis machinery. Keywords: Escherichia -
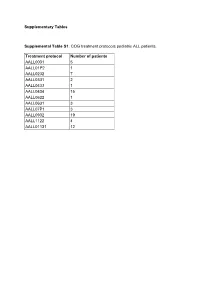
Supplementary Tables Supplemental Table S1. COG Treatment Protocols
Supplementary Tables Supplemental Table S1. COG treatment protocols pediatric ALL patients. Treatment protocol Number of patients AALL0031 5 AALL01P2 1 AALL0232 7 AALL0331 2 AALL0433 1 AALL0434 15 AALL0622 1 AALL0631 3 AALL07P1 3 AALL0932 19 AALL1122 4 AALL01131 12 Supplementary Table S2. This table contains the “Rosetta Stone” for the antibody and protein nomenclature (HUGO, MiMI, GeneCards) together with the RPPA staining details, including the primary and secondary antibody dilutions, the R2 scores For antibody validation, catalog number and the antibody source. Functional Protein Name Rosetta Stone RPPA Staining Details Category Functional Effect Common Name RPPA Antibody Huge Name (added MiMI Antibody R2, WB vs. Antibody 2nd Ab Full Name/Description from GeneCards of Mfg Claimed Manufacturer Catalog# Functional Group Name with PTMs) Name Source RPPA Dilution dilution Phosphorylation Target AKT1 AKT1 AKT1 v-akt murine thymoma viral oncogene homolog 1 AKT1 Cell Signaling 9272 Rabbit > 0.7 200 15000 PI3KAKT AKT1/AKT2/AKT3 AKT1_2_3.pS473 AKT1 v-akt murine thymoma viral oncogene homolog 1 Activation AKT-P473(Ser) Cell Signaling 9271 Rabbit > 0.7 150 20000 PI3KAKT Phospho Ser473 AKT1/AKT2/AKT3 AKT1_2_3.pT308 AKT1 v-akt murine thymoma viral oncogene homolog 1 Activation AKT-P308(Thr) Cell Signaling 9275 Rabbit > 0.7 200 20000 PI3KAKT Phospho Thr308 ASNS ASNS ASNS asparagine synthetase (glutamine-hydrolyzing) ASNS Sigma HPA029318 Rabbit 0.5-0.7 800 20000 Metabolic ATF3 ATF3 ATF3 activating transcription factor 3 ATF3 Abcam ab87213 Rabbit -

TAF4 Takes Flight
COMMENTARY TAF4 takes flight Michael T. Marr II1 Department of Biology and Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, MA 02454 he eukaryotic mRNA transcrip- from yeast to man, but the region of ETO-TAFH domain was required for tion machinery is exceedingly conservation is limited to a histone H2A activation of the wingless target gene complex, perhaps reflecting the homology region at the carboxyl termi- naked cuticle (nkd) both in cultured requirement for response to nus of the protein (Fig. 1A). The meta- cells and in larval imaginal discs. They Tdiverse environmental and developmen- zoan homologues of TAF4 contain an further showed that this domain directly tal signals. Some of the machinery is extended amino terminus with two addi- interacts with the amino terminus of the common to all mRNA genes and in- tional conserved regions: a glutamine- pygopus, a dedicated transcription factor cludes RNA polymerase II (Pol II) and rich region and a more recently defined for wingless signaling. Interestingly, the a set of general transcription factors ETO-TAFH domain. amino terminus of pygopus has previ- (GTFs) (1). Signal-specific gene expres- The glutamine-rich region was one of ously been defined as absolutely neces- sion patterns are defined by sequence- the first coactivator domains of TFIID sary for Wg/Wnt signaling (14). These specific DNA-binding proteins (activa- to be defined (8). It interacts with a findings place TAF4 at the end of a tors) that bind cognate sites in the number of activators including Sp1 and chain of protein–protein interactions promoter and enhancers of target genes.