10Th Anniversary International Multiconference BGRS\SB-2016
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

RESULTS Long Jump Women - Heptathlon
Moscow (RUS) World Championships 10-18 August 2013 RESULTS Long Jump Women - Heptathlon RESULT NAME COUNTRY AGE DATE VENUE World Record 7291 Jackie JOYNER-KERSEE USA 26 24 Sep 1988 Seoul Championships Record 7128 Jackie JOYNER-KERSEE USA 25 1 Sep 1987 Roma World Leading 6623 Tatyana CHERNOVA RUS 25 8 Jul 2013 Kazan TEMPERATURE HUMIDITY START TIME 09:30 16° C 88 % Group A 13 August 2013 END TIME 10:24 19° C 77 % PLACE BIB NAME COUNTRY DATE of BIRTH ORDER RESULT POINTS 1 2 3 1 432 Claudia RATH GER 25 Apr 86 16 6.67 PB 6.67 6.44 X 1.0 m/s 1.0 m/s 0.4 m/s 1.0 m/s Клаудиа Рат 25 апр . 86 1062 2 391 Katarina JOHNSON-THOMPSON GBR 09 Jan 93 15 6.56 PB X 6.56 X 0.6 m/s 1.1 m/s 0.6 m/s 0.8 m/s Катарина Джонсон -Томпсон 09 янв . 93 1027 3 856 Ganna MELNICHENKO UKR 24 Apr 83 12 6.49 SB 6.23 6.49 X 0.2 m/s 0.3 m/s 0.2 m/s 0.3 m/s Анна Мельниченко 24 апр . 83 1004 4 203 Brianne THEISEN EATON CAN 18 Dec 88 8 6.37 PB X 6.37 X 0.5 m/s 1.0 m/s 0.5 m/s 1.3 m/s Брайанн Тайсен Итон 18 дек . 88 965 5 624 Dafne SCHIPPERS NED 15 Jun 92 14 6.35 6.35 6.14 6.14 1.3 m/s 1.3 m/s 0.6 m/s 0.7 m/s Дафна Шипперс 15 июня 92 959 6 672 Karolina TYMINSKA POL 04 Oct 84 13 6.32 6.32 X 6.16 0.9 m/s 0.9 m/s 0.7 m/s 0.7 m/s Каролина Тыминская 04 окт . -

START LIST 100 Metres Hurdles Women - Heptathlon
REVISED Moscow (RUS) World Championships 10-18 August 2013 START LIST 100 Metres Hurdles Women - Heptathlon CONFIRMATION OF ATHLETES 377 MADE IN ERROR. REMOVE FROM START LIST. RESULT NAME COUNTRY AGE DATE VENUE World Record 7291 Jackie JOYNER-KERSEE USA 26 24 Sep 1988 Seoul Championships Record 7128 Jackie JOYNER-KERSEE USA 25 1 Sep 1987 Roma World Leading 6623 Tatyana CHERNOVA RUS 25 8 Jul 2013 Kazan i = Indoor performance Heat 1 12 August 2013 9:30 4 LANE BIB NAME COUNTRY DATE of BIRTH PERSONAL BEST SEASON BEST 2 321 Mari KLAUP EST 27 Feb 90 14.09 14.09 Мари Клауп 27 февр . 90 3 423 Julia MÄCHTIG GER 01 Jan 86 14.10 14.12 Юлия Мехтих 01 янв . 86 4 453 Györgyi FARKAS-ZSIVOCZKY HUN 13 Feb 85 13.97 13.97 Дьордьи Фаркаш -Живоцки 13 февр . 85 5 821 Wassana WINATHO THA 30 Jun 80 13.62 13.90 Вассана Уинато 30 июня 80 6 293 Eliška KLUCINOVÁ CZE 14 Apr 88 14.01 14.06 Элишка Клюцинова 14 апр . 88 7 640 Ida MARCUSSEN NOR 01 Nov 87 13.96 14.11 Ида Маркуссен 01 нояб . 87 8 713 Aleksandra BUTVINA RUS 14 Feb 86 14.04 14.04 Александра Бутвина 14 февр . 86 9 545 Irina KARPOVA KAZ 13 Feb 80 13.96 14.52 Ирина Карпова 13 февр . 80 Heat 2 12 August 2013 9:38 4 LANE BIB NAME COUNTRY DATE of BIRTH PERSONAL BEST SEASON BEST 2 163 Katsiaryna NETSVIATAYEVA BLR 28 Jun 89 13.74 13.84 Катерина Нецвятаева 28 июня 89 3 804 Linda ZÜBLIN SUI 21 Mar 86 13.55 13.76 Линда Зюблин 21 марта 86 4 148 Nafissatou THIAM BEL 19 Aug 94 13.87 13.87 Нафиссату Тиам 19 авг . -

BGRS\SB-2018 MM&Am
11th International Multiconference «Bioinformatics of Genome Regulation and Structure\ Systems Biology», Novosibirsk, Russia, 20 - 25 August 2018 BGRS\SB-2018 MM&HPC-BBB-2018 SBioMed-2018 CSGB-2018 BioGenEvo-2018 SbPCD-2018 FCRW-2018 20 August 08:30-10:00 Registration (House of Scientists SB RAS, main entrance) Coffee-break 10.10–17.30 Plenary session (House of Scientists SB RAS, Large hall) Chairpersons: Prof. Nikolay Kolchanov, Prof. Ralf Hofestädt, Prof. Mikhail Fedoruk 10.10-10.30 Opening Ceremony (House of Scientists SB RAS, Large Hall) 10.30–11.10 Entropic hourglass patterns of animal and plant development and the emergence of biodiversity. Ivo Grosse, Institute of Computer Science, Martin Luther University Halle-Wittenberg, Halle, Germany 11.10–11.50 Selection versus Adaptation: Network Diversification and the Origins of Life, Ageing and Cancer Hans V. Westerhoff, Synthetic Systems Biology and Nuclear Organization, University of Amsterdam 11.50–12.30 Unraveling of gene expression control in genome-reduced bacteria. The rally goes on… Vadim Govorun, Federal Research and Clinical Center of Physical-Chemical Medicine of Federal Medical Biological Agency (RCC PCM FMBA), Russia 12.30–12.45 HPC clusters and Big Data storage for data analysis in scientific research – Huawei experience Alexey Iyudin, Huawei Technologies, Gold Sponsor 12.45–14.30 Lunch + Registration 14.30–15.10 Towards understanding of apoptosis regulation using computational biology Inna N. Lavrik, Department of Translational Inflammation Research, Institute of Experimental -

London 2012 Athletics Day 1 Results
http://www.iaaf.org/results/eventCode=4871/raceDate=08-03-2012/txt... ------------------------------------------------------------------------------------- The XXX Olympic Games London (OS) (GBR) - Friday, Aug 03, 2012 Shot Put - M QUALIFICATION Qual. rule: qualification standard 20.65m or at least best 12 qualified. ------------------------------------------------------------------------------------- Group A 03 August 2012 - 10:00 Position Bib Athlete Country Mark . 1 1924 David Storl GER 21.15 Q . 2 2659 Tomasz Majewski POL 21.03 Q . 3 1023 Germán Lauro ARG 20.75 Q (NR) 4 1163 Pavel Lyzhyn BLR 20.57 q (SB) 5 3209 Christian Cantwell USA 20.41 q . 6 2187 Dorian Scott JAM 20.31 q . 7 2836 Soslan Tsirikhov RUS 20.17 . 8 2550 Rutger Smith NED 20.08 . 9 2711 Marco Fortes POR 20.06 . 10 2046 Lajos Kürthy HUN 19.65 . 11 1264 Georgi Ivanov BUL 19.63 . 12 1153 Kemal Mesic BIH 19.60 . 13 1986 Mihaíl Stamatóyiannis GRE 19.24 . 14 1063 Dale Stevenson AUS 19.17 . 15 2395 Maris Urtans LAT 19.13 . 16 1639 Borja Vivas ESP 18.88 . 17 2505 Stephen Saenz MEX 18.65 . 18 2118 Ódinn Björn Thorsteinsson ISL 17.62 . 19 1003 Adriatik Hoxha ALB 17.58 . 1297 Justin Rodhe CAN NM . Athlete 1st 2nd 3rd David Storl 21.15 Tomasz Majewski 21.03 Germán Lauro 20.10 X 20.75 Pavel Lyzhyn 19.85 20.10 20.57 Christian Cantwell 20.41 20.30 19.88 Dorian Scott 20.18 20.30 20.31 Soslan Tsirikhov 19.25 19.78 20.17 Rutger Smith 20.08 X 19.55 Marco Fortes 19.59 20.06 19.50 Lajos Kürthy 19.65 X X Georgi Ivanov 19.42 19.40 19.63 Kemal Mesic 19.49 X 19.60 Mihaíl Stamatóyiannis 18.67 X 19.24 Dale Stevenson 18.38 19.17 19.01 Maris Urtans 18.92 19.13 X Borja Vivas 18.46 X 18.88 Stephen Saenz X 18.65 X Ódinn Björn Thorsteinsson X 17.04 17.62 Adriatik Hoxha 17.58 X 17.13 Justin Rodhe X X X Group B 03 August 2012 - 10:00 Position Bib Athlete Country Mark . -
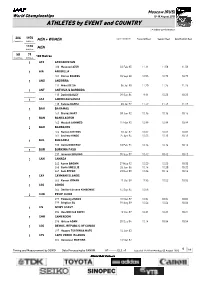
0 E Event Country
Moscow (RUS) World Championships 10-18 August 2013 ATHLETES by EVENT and COUNTRY i = Indoor performance 206 1970 MEN + WOMEN DATE of BIRTH Personal Best Season Best Qualification Best Countries Athletes 1105 MEN Athletes 58 79 100 Metres Countries Athletes 1 AFG AFGHANISTAN 100 Massoud AZIZI 02 Feb 85 11.11 11.58 11.58 1 AIA ANGUILLA 101 Kieron ROGERS 08 Sep 88 10.55 10.75 10.75 1 AND ANDORRA 111 Mikel DE SA 06 Jul 90 11.20 11.26 11.26 1 ANT ANTIGUA & BARBUDA 113 Daniel BAILEY 09 Sep 86 9.91 10.23 10.23 1 ASA AMERICAN SAMOA 118 Faresa KAPISI 23 Jul 97 11.47 11.47 11.47 1 BAH BAHAMAS 161 Shavez HART 09 Jun 92 10.16 10.16 10.16 1 BAN BANGLADESH 165 Masbah AHMMED 11 Mar 95 10.99 10.99 10.99 2 BAR BARBADOS 166 Ramon GITTENS 20 Jul 87 10.02 10.02 10.02 167 Andrew HINDS 25 Apr 84 10.03 10.13 10.13 1 BUL BULGARIA 233 Denis DIMITROV 10 Feb 94 10.16 10.16 10.16 1 BUR BURKINA FASO 237 Innocent BOLOGO 05 Sep 89 10.42 10.42 10.42 3 CAN CANADA 243 Aaron BROWN 27 May 92 10.05 10.05 10.05 245 Gavin SMELLIE 26 Jun 86 10.14 10.20 10.20 247 Sam EFFAH 29 Dec 88 10.06 10.14 10.14 1 CAY CAYMAN ISLANDS 263 Kemar HYMAN 11 Oct 89 9.95 10.02 10.02 1 CGO CONGO 264 Devilert Arsene KIMBEMBE 14 Sep 84 10.58 2 CHN PR OF CHINA 277 Peimeng ZHANG 13 Mar 87 10.04 10.04 10.04 279 Bingtian SU 29 Aug 89 10.06 10.06 10.06 1 CIV IVORY COAST 294 Hua Wilfried KOFFI 12 Oct 87 10.21 10.21 10.21 1 CMR CAMEROON 296 Idrissa ADAM 28 Dec 84 10.14 10.54 10.54 1 COD DEMOC. -

20Th Asian Athletics Championships
20th Asian Athletics Championships-2013 * 03rd to 07th July 2013 * Pune, Maharastra, INDIA * *~*~* Final Results *~*~* Event # 1 MENS 100m Time:-19:15 Hrs 04/07/2013 # W.R.:-:USAIN BOLT JAM,BERLIN,GER 9.58 16-8-2009 % A.R.:-:Samuel Francis QAT;Amman 9.99 26.07.2007 &M.R.:-:S.Francis QAT; Amman 9.99 26.07.2007 Final 1 SU BINGTIAN 29/08/89 CHN -0.3m/s 10.17 2 SAMUEL ADELEBARI FRANCIS 27/03/87 QAT -0.3m/s 10.27 3 BARAKAT AL HARTHI 15/06/88 OMA -0.3m/s 10.30 4 REZA GHASEMI 24/07/87 IRI -0.3m/s 10.37 5 FAHAD MOHAMMED ALSUBAIE 04/02/94 KSA -0.3m/s 10.48 6 NG KA FUNG 27/10/92 HKG -0.3m/s 10.52 7 NAOKI TSUKAHARA 10/05/85 JPN -0.3m/s 10.54 8 HASSAN TAFTIAN 04/05/93 IRI -0.3m/s 10.54 Semi Final-1 1 SU BINGTIAN 29/08/89 CHN -1.1M/S 10.37 2 REZA GHASEMI 24/07/87 IRI -1.1M/S 10.38 3 FAHAD MOHAMMED ALSUBAIE 04/02/94 KSA -1.1M/S 10.48 4 TSUI CHI HO 17/02/90 HKG -1.1M/S 10.52 5 ANIRUDDH KALIDAS GUJJAR 14/11/90 IND -1.1M/S 10.59 6 MOHAMED SALMAN 04/04/92 BRN -1.1M/S 10.86 EID ABDULLA E A AL KUWARI 04/02/93 QAT DQ SOTA KAWATSURA 19/06/89 JPN DQ Semi Final-2 1 BARAKAT AL HARTHI 15/06/88 OMA -0.5M/S 10.27 2 SAMUEL ADELEBARI FRANCIS 27/03/87 QAT -0.5M/S 10.30 3 HASSAN TAFTIAN 04/05/93 IRI -0.5M/S 10.32 4 NAOKI TSUKAHARA 10/05/85 JPN -0.5M/S 10.44 5 NG KA FUNG 27/10/92 HKG -0.5M/S 10.47 6 ALI AL DOSERI 02/03/83 BRN -0.5M/S 10.59 7 LIAQAT ALI 06/08/83 PAK -0.5M/S 10.64 8 SAPWATUR RAHMAN 13/05/94 INA -0.5M/S 10.78 Heat-1 1 REZA GHASEMI 24/07/87 IRI -0.2m/s 10.41 2 NG KA FUNG 27/10/92 HKG -0.2m/s 10.51 3 ALI AL DOSERI 02/03/83 BRN -0.2m/s 10.72 4 KRITSADA NAMSUWAN -

ETN2012 53(Wogii)
Volume 11, No. 53 August 17, 2012 version ii — Olympic Games Women — — By–Nation Medal Chart — LONDON, ENGLAND AUGUST 3–12 Nation ................Men Women Overall G S B Total G S B Total ATTENDANCE United States ......3 9 3 15 6 4 4 14 ........29 No official figures released, but all sessions should be considered Russia .................2 — 2 6 5 5 16 ........18 “80,000-seat sellouts.” Jamaica ..............3 2 2 7 1 2 2 5 ..........12 WEATHER Kenya ..................2 1 4 7 — 3 1 4 ..........11 Official temperature (both F and C) and humidity readings are given Germany .............1 2 1 4 — 2 2 4 ............8 with each event. Spotty rain on several days; generally pleasant Ethiopia ...............— 1 1 2 3 — 2 5 ............7 (c70F/20C) and calm. In almost all cases the readings are for the China ..................1 — 2 3 — — 3 3 ............6 beginning of the event. Great Britain ........3 — 1 4 1 1 — 2 ............6 Trinidad ...............1 — 3 4 — — — — ...........4 EXPLAINING THE TYPOGRAPHY Paragraph breaks in the preliminary rounds of running events indicate Australia ..............— 2 — 2 1 — — 1 ............3 the separation between qualifiers and non-qualifiers. Ukraine ...............— 1 — 1 — — 2 2 ............3 Cuba ...................— 1 1 — 1 — 1 ............2 COLOR CODING Czech Republic...— — — 1 — 1 2 ............2 All medalists appear in purple ink; all Americans are underlined (if Dominican Rep ...1 1 — 2 — — — — ...........2 in multiple rounds, only in the final round in which they competed); France .................1 1 — 2 — — — — ...........2 field-event/multi medalists appear in either blue (gold medal), red Poland ................1 — 1 — 1 — 1 ............2 (silver) or green (bronze) in the progression charts. -
Asian Athletics Digest February 15, 2016 4 / 2016
Asian Athletics Digest February 15, 2016 4 / 2016 Asian Indoor Championships Special Preview ~ Ranklists ~ Records Asian Athletics Digest February 15, 2016 Contents From the Editor’s desk .……………… 66 Preview on Asian Indoor Champs …. 67 From the Editor’s desk Asian Indoor All-Time Rankings ……. 68 Asian & National Indoor Records ….. 75 Hello All, Welcome back to Asian Athletics Digest! First of all I would like to thank you all for your No. 4/2016 comments, continuous support and encouragement to our magazine from time to time. Editor & Publisher Ram. Murali Krishnan This edition of our magazine is dedicated to the 7th Asian Indoor Athletics Championships to be held Doha, Qatar, from 19 Address for Communication to 20 February 2016. “ Kausalyam ” 72/4, Sri Padmavathi Nagar NSN Palayam P.O. As in the previous edition, that was produced as a Coimbatore – 641031, INDIA special preview for the South Asian Games, this number of our magazine will also have tons of stats on Asian Indoor Athletics Contact E-Mail [email protected] As much as 432 athletes from 37 member national [email protected] federations of Asian Athletics Association are expected to take part in the three day meet at Doha. PHOTOS Doha 2016 Organising Committee In this edition of Asian Athletics Digest contains a brief history of Asian Indoor championships, Asian All-Time Top-10 Designer ranking lists as well as Asian and National Indoor Records. N. Sasivarnan To accommodate the large number of info in this issue we have excluded some of our regular features, which will be resumed in our next number. -

Wyniki / • REZULTATY
Men 100 m 5 sierpniaWind: 1.5 1 Usain Bolt JAM 9.63 SB WL 0.165 2 Yohan Blake JAM 9.75 0.179 3 Justin Gatlin USA 9.79 PB 0.178 4 Tyson Gay USA 9.80 SB 0.145 5 Ryan Bailey USA 9.88 0.176 6 Churandy Martina NED 9.94 0.139 7 Richard Thompson TRI 9.98 0.160 8 Asafa Powell JAM 11.99 0.155 Semifinals 5 sierpnia Heat 1 Wind: 0.7 1 Justin Gatlin USA 9.82 Q 0.187 2 Churandy Martina NED 9.91 NR PB Q 0.148 3 Asafa Powell JAM 9.94 q 0.155 4 Keston Bledman TRI 10.04 0.175 5 Ben Youssef Meité CIV 10.13 0.163 6 Jimmy Vicaut FRA 10.16 0.203 7 James Dasaolu GBR 10.18 0.174 8 Su Wai'bou Sanneh GAM 10.18 NR PB 0.175 Heat 2 Wind: 1.0 1 Usain Bolt JAM 9.87 Q 0.180 2 Ryan Bailey USA 9.96 Q 0.155 3 Richard Thompson TRI 10.02 q 0.158 4 Dwain Chambers GBR 10.05 0.154 5 Gerald Phiri ZAM 10.11 SB 0.165 6 Daniel Bailey ANT 10.16 0.142 7 Antoine Adams SKN 10.27 0.159 8 Su Bingtian CHN 10.28 0.157 Heat 3 Wind: 1.7 1 Yohan Blake JAM 9.85 Q 0.176 2 Tyson Gay USA 9.90 Q 0.151 3 Adam Gemili GBR 10.06 0.158 4 Derrick Atkins BAH 10.08 SB 0.164 5 Justyn Warner CAN 10.09 0.135 6 Ryota Yamagata JPN 10.10 0.158 7 Rondell Sorrillo TRI 10.31 0.140 Kemar Hyman CAY DNS Heats 4 sierpnia Heat 1 Wind: -1.4 1 Tyson Gay USA 10.08 Q 0.147 2 Richard Thompson TRI 10.14 Q 0.151 3 Gerald Phiri ZAM 10.16 Q 0.147 4 Jaysuma Saidy Ndure NOR 10.28 0.166 5 Ángel David Rodríguez ESP 10.34 0.168 6 Jurgen Themen SUR 10.53 0.169 7 Isidro Montoya COL 10.54 0.165 8 Gary Yeo Foo Ee SIN 10.69 0.144 Heat 2 Wind: 0.7 1 Justin Gatlin USA 9.97 Q 0.200 2 Derrick Atkins BAH 10.22 Q 0.179 3 Rondell Sorrillo -
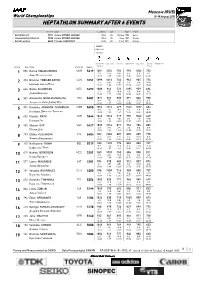
Heptathlon Summary After 6 Events
Moscow (RUS) World Championships 10-18 August 2013 HEPTATHLON SUMMARY AFTER 6 EVENTS RESULT NAME COUNTRY AGE DATE VENUE World Record 7291 Jackie JOYNER-KERSEE USA 26 24 Sep 1988 Seoul Championships Record 7128 Jackie JOYNER-KERSEE USA 25 1 Sep 1987 Roma World Leading 6623 Tatyana CHERNOVA RUS 25 8 Jul 2013 Kazan POINTS RANK-HEAT RESULT 100 Metres High Jump Shot Put 200 Metres Long Jump Javelin 800 Metres Hurdles Throw PLACE BIB NAME COUNTRY RESULT 856 Ganna MELNICHENKO UKR 5619 1081 1054 784 993 1004 703 1 2-4 5-A 8-A 4-5 3-A 1-B Анна Мельниченко 13.29 1.86 13.85 23.87 6.49 41.87 203 Brianne THEISEN EATON CAN 5551 1099 1016 732 963 965 776 2 1-4 6-A 1-B 1-4 4-A 10-A Брайанн Тайсен Итон 13.17 1.83 13.07 24.18 6.37 45.64 624 Dafne SCHIPPERS NED 5492 1080 941 721 1095 959 696 3 3-4 6-B 16-A 1-5 5-A 3-B Дафна Шипперс 13.30 1.77 12.91 22.84 6.35 41.47 369 Antoinette NANA DJIMOU IDA FRA 5481 1071 941 830 891 840 908 4 4-4 7-B 2-A 3-3 15-A 2-A Антуанетт Нана Джиму Ида 13.36 1.77 14.54 24.95 5.97 52.47 391 Katarina JOHNSON-THOMPSON GBR 5450 1052 1016 629 1042 1027 684 5 6-4 7-A 14-B 2-5 2-A 4-B Катарина Джонсон -Томпсон 13.49 1.83 11.52 23.37 6.56 40.86 432 Claudia RATH GER 5444 1043 1016 719 955 1062 649 6 7-4 1-B 4-B 2-4 1-A 10-B Клаудиа Рат 13.55 1.83 12.88 24.27 6.67 39.04 900 Sharon DAY USA 5427 1049 1016 817 954 786 805 7 2-3 8-A 3-A 3-4 9-B 8-A Шэрон Дэй 13.51 1.83 14.35 24.28 5.79 47.17 293 Eliška KLUCINOVÁ CZE 5404 983 1054 807 895 887 778 8 2-1 4-A 5-A 2-3 10-A 9-A Элишка Клюцинова 13.97 1.86 14.19 24.91 6.12 45.76 148 Nafissatou THIAM -

RESULTS 400 Metres Men - Final
Moscow (RUS) World Championships 10-18 August 2013 RESULTS 400 Metres Men - Final RESULT NAME COUNTRY AGE DATE VENUE World Record 43.18 Michael JOHNSON USA 32 26 Aug 1999 Sevilla Championships Record 43.18 Michael JOHNSON USA 32 26 Aug 1999 Sevilla World Leading 43.74 LaShawn MERRITT USA 27 13 Aug 2013 Moscow TEMPERATURE HUMIDITY START TIME 21:49 24° C 47 % Final 13 August 2013 PLACE BIB NAME COUNTRY DATE of BIRTH LANE RESULT REACTION Fn 1 1150 LaShawn MERRITT USA 27 Jun 86 6 43.74 WL 0.256 ЛаШон Мерритт 27 июня 86 2 1148 Tony MCQUAY USA 16 Apr 90 4 44.40 PB 0.155 Тони МакКвей 16 апр . 90 3 349 Luguelín SANTOS DOM 12 Nov 93 7 44.52 SB 0.350 Лугелин Сантос 12 нояб . 93 4 176 Jonathan BORLÉE BEL 22 Feb 88 8 44.54 SB 0.224 Джонатан Борле 22 февр . 88 5 330 Pavel MASLÁK CZE 21 Feb 91 1 44.91 0.169 Павел Маслак 21 февр . 91 6 744 Yousef Ahmed MASRAHI KSA 31 Dec 87 3 44.97 0.162 Юсеф Ахмед Масрахи 31 дек . 87 7 560 Kirani JAMES GRN 01 Sep 92 5 44.99 0.186 Кирани Джеймс 01 сент . 92 8 210 Anderson HENRIQUES BRA 03 Mar 92 2 45.03 0.157 Андерсон Энрикес 03 марта 92 ALL-TIME TOP LIST SEASON TOP LIST RESULT NAME Venue DATE RESULT NAME Venue DATE 43.18 Michael JOHNSON (USA) Sevilla 26 Aug 99 43.74 LaShawn MERRITT (USA) Moscow 13 Aug 13 43.29 Harry REYNOLDS (USA) Zürich (Letzigrund) 17 Aug 88 43.96 Kirani JAMES (GRN) Paris Saint-Denis 6 Jul 13 43.45 Jeremy WARINER (USA) Osaka 31 Aug 07 44.40 Tony MCQUAY (USA) Moscow 13 Aug 13 43.50 Quincy WATTS (USA) Barcelona 5 Aug 92 44.52 Luguelín SANTOS (DOM) Moscow 13 Aug 13 43.74 LaShawn MERRITT (USA) Moscow -

Prospectus 2010-11
University Pathways 2010-2011 ACADEMIC AND ENGLISH LANGUAGE PREPARATION FOR INTERNATIONAL STUDENTS g INTO UNIVERSITY OF EXETER YOUR BEST ROUTE TO UNIVERSITY SUCCESS 2010 rankings Study in one of the most beautiful campuses in Britain and live in the safe and student-friendly city of Exeter. • 9th in The Times Good University Guide 2010 • 5th in the 2009 National Student Survey for overall student satisfaction • 21 subjects ranked in the top 10 in the UK including General Engineering, Sports Science, Economics, Drama, Politics, Psychology and Middle East Studies* • 2nd in The Times for Accounting and Finance, 5th for Business Studies and 6th for Economics* • Law School ranks 6th in the UK for overall satisfaction (2009 NSS) • 90 per cent of research is internationally recognised (RAE 2008) * Times Good University Guide 2010 Welcome A welcome message from A welcome message Contents the Vice-Chancellor from INTO University of Exeter 4 International students studying at the INTO INTO University of Exeter has just completed University of Exeter Centre have a strong track another highly successful year. Nearly 300 Academic, cultural and social facilities 8 record of success. Results from our latest cohort international students progressed to the The INTO Centre 10 indicate that over 95 per cent of students passed University from our academic preparation Exeter and the South West 12 the Foundation programme and every one programmes, and another 400 students entered Choosing an INTO course 14 of the Graduate Certificate students passed. directly from our English language courses. Congratulations to all of them! We are also delighted The number of students choosing to study at Progress and outcomes 17 to announce that over 90 per cent of students the Centre continues to rise, supported by Foundation programme 18 completing academic preparation programmes who the top-ten status of the University, the good Foundation module descriptions 24 were eligible to progress onto one of the University’s reputation of our courses, the excellent student degrees have chosen to do so.