Supplemental Information Complement Protein C1q Directs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Connexin 40.1 (GJD4) (NM 153368) Human Tagged ORF Clone Lentiviral Particle – RC222438L3V | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC222438L3V Connexin 40.1 (GJD4) (NM_153368) Human Tagged ORF Clone Lentiviral Particle Product data: Product Type: Lentiviral Particles Product Name: Connexin 40.1 (GJD4) (NM_153368) Human Tagged ORF Clone Lentiviral Particle Symbol: GJD4 Synonyms: CX40.1 Vector: pLenti-C-Myc-DDK-P2A-Puro (PS100092) ACCN: NM_153368 ORF Size: 1110 bp ORF Nucleotide The ORF insert of this clone is exactly the same as(RC222438). Sequence: OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_153368.1 RefSeq Size: 1580 bp RefSeq ORF: 1113 bp Locus ID: 219770 UniProt ID: Q96KN9 Protein Families: Transmembrane MW: 40 kDa Gene Summary: Connexins, such as GJD4, are involved in the formation of gap junctions, intercellular conduits that directly connect the cytoplasms of contacting cells. Each gap junction channel is formed by docking of 2 hemichannels, each of which contains 6 connexin subunits (Sohl et al., 2003 [PubMed 12881038]).[supplied by OMIM, Mar 2008] This product is to be used for laboratory only. -

The Significance of the Evolutionary Relationship of Prion Proteins and ZIP Transporters in Health and Disease
The Significance of the Evolutionary Relationship of Prion Proteins and ZIP Transporters in Health and Disease by Sepehr Ehsani A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Department of Laboratory Medicine and Pathobiology University of Toronto © Copyright by Sepehr Ehsani 2012 The Significance of the Evolutionary Relationship of Prion Proteins and ZIP Transporters in Health and Disease Sepehr Ehsani Doctor of Philosophy Department of Laboratory Medicine and Pathobiology University of Toronto 2012 Abstract The cellular prion protein (PrPC) is unique amongst mammalian proteins in that it not only has the capacity to aggregate (in the form of scrapie PrP; PrPSc) and cause neuronal degeneration, but can also act as an independent vector for the transmission of disease from one individual to another of the same or, in some instances, other species. Since the discovery of PrPC nearly thirty years ago, two salient questions have remained largely unanswered, namely, (i) what is the normal function of the cellular protein in the central nervous system, and (ii) what is/are the factor(s) involved in the misfolding of PrPC into PrPSc? To shed light on aspects of these questions, we undertook a discovery-based interactome investigation of PrPC in mouse neuroblastoma cells (Chapter 2), and among the candidate interactors, identified two members of the ZIP family of zinc transporters (ZIP6 and ZIP10) as possessing a PrP-like domain. Detailed analyses revealed that the LIV-1 subfamily of ZIP transporters (to which ZIPs 6 and 10 belong) are in fact the evolutionary ancestors of prions (Chapter 3). -

C1orf21 CRISPR/Cas9 KO Plasmid (H): Sc-417269
SANTA CRUZ BIOTECHNOLOGY, INC. C1orf21 CRISPR/Cas9 KO Plasmid (h): sc-417269 BACKGROUND APPLICATIONS The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and C1orf21 CRISPR/Cas9 KO Plasmid (h) is recommended for the disruption of CRISPR-associated protein (Cas9) system is an adaptive immune response gene expression in human cells. defense mechanism used by archea and bacteria for the degradation of foreign genetic material (4,6). This mechanism can be repurposed for other 20 nt non-coding RNA sequence: guides Cas9 functions, including genomic engineering for mammalian systems, such as to a specific target location in the genomic DNA gene knockout (KO) (1,2,3,5). CRISPR/Cas9 KO Plasmid products enable the U6 promoter: drives gRNA scaffold: helps Cas9 identification and cleavage of specific genes by utilizing guide RNA (gRNA) expression of gRNA bind to target DNA sequences derived from the Genome-scale CRISPR Knock-Out (GeCKO) v2 library developed in the Zhang Laboratory at the Broad Institute (3,5). Termination signal Green Fluorescent Protein: to visually REFERENCES verify transfection CRISPR/Cas9 Knockout Plasmid CBh (chicken β-Actin 1. Cong, L., et al. 2013. Multiplex genome engineering using CRISPR/Cas hybrid) promoter: drives expression of Cas9 systems. Science 339: 819-823. 2A peptide: allows production of both Cas9 and GFP from the 2. Mali, P., et al. 2013. RNA-guided human genome engineering via Cas9. same CBh promoter Science 339: 823-826. Nuclear localization signal 3. Ran, F.A., et al. 2013. Genome engineering using the CRISPR-Cas9 system. Nuclear localization signal SpCas9 ribonuclease Nat. Protoc. 8: 2281-2308. -

Podocyte Specific Knockdown of Klf15 in Podocin-Cre Klf15flox/Flox Mice Was Confirmed
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1: Podocyte specific knockdown of Klf15 in Podocin-Cre Klf15flox/flox mice was confirmed. (A) Primary glomerular epithelial cells (PGECs) were isolated from 12-week old Podocin-Cre Klf15flox/flox and Podocin-Cre Klf15+/+ mice and cultured at 37°C for 1 week. Real-time PCR was performed for Nephrin, Podocin, Synaptopodin, and Wt1 mRNA expression (n=6, ***p<0.001, Mann-Whitney test). (B) Real- time PCR was performed for Klf15 mRNA expression (n=6, *p<0.05, Mann-Whitney test). (C) Protein was also extracted and western blot analysis for Klf15 was performed. The representative blot of three independent experiments is shown in the top panel. The bottom panel shows the quantification of Klf15 by densitometry (n=3, *p<0.05, Mann-Whitney test). (D) Immunofluorescence staining for Klf15 and Wt1 was performed in 12-week old Podocin-Cre Klf15flox/flox and Podocin-Cre Klf15+/+ mice. Representative images from four mice in each group are shown in the left panel (X 20). Arrows show colocalization of Klf15 and Wt1. Arrowheads show a lack of colocalization. Asterisk demonstrates nonspecific Wt1 staining. “R” represents autofluorescence from RBCs. In the right panel, a total of 30 glomeruli were selected in each mouse and quantification of Klf15 staining in the podocytes was determined by the ratio of Klf15+ and Wt1+ cells to Wt1+ cells (n=6 mice, **p<0.01, unpaired t test). Supplementary Figure 2: LPS treated Podocin-Cre Klf15flox/flox mice exhibit a lack of recovery in proteinaceous casts and tubular dilatation after DEX administration. -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

Ankrd9 Is a Metabolically-Controlled Regulator of Impdh2 Abundance and Macro-Assembly
ANKRD9 IS A METABOLICALLY-CONTROLLED REGULATOR OF IMPDH2 ABUNDANCE AND MACRO-ASSEMBLY by Dawn Hayward A dissertation submitted to The Johns Hopkins University in conformity with the requirements of the degree of Doctor of Philosophy Baltimore, Maryland April 2019 ABSTRACT Members of a large family of Ankyrin Repeat Domains proteins (ANKRD) regulate numerous cellular processes by binding and changing properties of specific protein targets. We show that interactions with a target protein and the functional outcomes can be markedly altered by cells’ metabolic state. ANKRD9 facilitates degradation of inosine monophosphate dehydrogenase 2 (IMPDH2), the rate-limiting enzyme in GTP biosynthesis. Under basal conditions ANKRD9 is largely segregated from the cytosolic IMPDH2 by binding to vesicles. Upon nutrient limitation, ANKRD9 loses association with vesicles and assembles with IMPDH2 into rod-like structures, in which IMPDH2 is stable. Inhibition of IMPDH2 with Ribavirin favors ANKRD9 binding to rods. The IMPDH2/ANKRD9 assembly is reversed by guanosine, which restores association of ANKRD9 with vesicles. The conserved Cys109Cys110 motif in ANKRD9 is required for the vesicles-to-rods transition as well as binding and regulation of IMPDH2. ANKRD9 knockdown increases IMPDH2 levels and prevents formation of IMPDH2 rods upon nutrient limitation. Thus, the status of guanosine pools affects the mode of ANKRD9 action towards IMPDH2. Advisor: Dr. Svetlana Lutsenko, Department of Physiology, Johns Hopkins University School of Medicine Second reader: -

REPORT Germline Mutation of INI1/SMARCB1 in Familial Schwannomatosis
REPORT Germline Mutation of INI1/SMARCB1 in Familial Schwannomatosis Theo J. M. Hulsebos, Astrid S. Plomp, Ruud A. Wolterman, Els C. Robanus-Maandag, Frank Baas, and Pieter Wesseling Patients with schwannomatosis develop multiple schwannomas but no vestibular schwannomas diagnostic of neurofi- bromatosis type 2. We report an inactivating germline mutation in exon 1 of the tumor-suppressor gene INI1 in a father and daughter who both had schwannomatosis. Inactivation of the wild-type INI1 allele, by a second mutation in exon 5 or by clear loss, was found in two of four investigated schwannomas from these patients. All four schwannomas displayed complete loss of nuclear INI1 protein expression in part of the cells. Although the exact oncogenetic mechanism in these schwannomas remains to be elucidated, our findings suggest that INI1 is the predisposing gene in familial schwannomatosis. Schwannomatosis (MIM 162091) is characterized by the 10 Only two families with an INI1 germline mutation, an development of multiple spinal, peripheral, and cranial- exon 4 frameshift mutation,11 and an exon 7 donor splice nerve schwannomas in the absence of vestibular schwan- site mutation12 have been described in which multiple nomas.1 The presence of vestibular schwannomas is di- generations were affected by malignant (rhabdoid) tumors agnostic of neurofibromatosis type 2 (NF2 [MIM 101000]). in infancy. In both of these families, clear cases of no- Molecular analyses identified somatically acquired mu- nexpressing obligate carriers of the INI1 mutation were -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Figure S1. Representative Report Generated by the Ion Torrent System Server for Each of the KCC71 Panel Analysis and Pcafusion Analysis
Figure S1. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. A Figure S1. Continued. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. B Figure S2. Comparative analysis of the variant frequency found by the KCC71 panel and calculated from publicly available cBioPortal datasets. For each of the 71 genes in the KCC71 panel, the frequency of variants was calculated as the variant number found in the examined cases. Datasets marked with different colors and sample numbers of prostate cancer are presented in the upper right. *Significantly high in the present study. Figure S3. Seven subnetworks extracted from each of seven public prostate cancer gene networks in TCNG (Table SVI). Blue dots represent genes that include initial seed genes (parent nodes), and parent‑child and child‑grandchild genes in the network. Graphical representation of node‑to‑node associations and subnetwork structures that differed among and were unique to each of the seven subnetworks. TCNG, The Cancer Network Galaxy. Figure S4. REVIGO tree map showing the predicted biological processes of prostate cancer in the Japanese. Each rectangle represents a biological function in terms of a Gene Ontology (GO) term, with the size adjusted to represent the P‑value of the GO term in the underlying GO term database. -

Repeat Proteins Challenge the Concept of Structural Domains
844 Biochemical Society Transactions (2015) Volume 43, part 5 Repeat proteins challenge the concept of structural domains Rocıo´ Espada*1, R. Gonzalo Parra*1, Manfred J. Sippl†, Thierry Mora‡, Aleksandra M. Walczak§ and Diego U. Ferreiro*2 *Protein Physiology Lab, Dep de Qu´ımica Biologica, ´ Facultad de Ciencias Exactas y Naturales, UBA-CONICET-IQUIBICEN, Buenos Aires, C1430EGA, Argentina †Center of Applied Molecular Engineering, Division of Bioinformatics, Department of Molecular Biology, University of Salzburg, 5020 Salzburg, Austria ‡Laboratoire de physique statistique, CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France §Laboratoire de physique theorique, ´ CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France Abstract Structural domains are believed to be modules within proteins that can fold and function independently. Some proteins show tandem repetitions of apparent modular structure that do not fold independently, but rather co-operate in stabilizing structural forms that comprise several repeat-units. For many natural repeat-proteins, it has been shown that weak energetic links between repeats lead to the breakdown of co-operativity and the appearance of folding sub-domains within an apparently regular repeat array. The quasi-1D architecture of repeat-proteins is crucial in detailing how the local energetic balances can modulate the folding dynamics of these proteins, which can be related to the physiological behaviour of these ubiquitous biological systems. Introduction and between repeats, challenging the concept of structural It was early on noted that many natural proteins typically domain. collapse stretches of amino acid chains into compact units, defining structural domains [1]. These domains typically correlate with biological activities and many modern proteins can be described as composed by novel ‘domain arrange- Definition of the repeat-units ments’ [2]. -

A Structural Guide to Proteins of the NF-Kb Signaling Module
Downloaded from http://cshperspectives.cshlp.org/ on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press A Structural Guide to Proteins of the NF-kB Signaling Module Tom Huxford1 and Gourisankar Ghosh2 1Department of Chemistry and Biochemistry, San Diego State University, 5500 Campanile Drive, San Diego, California 92182-1030 2Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92093-0375 Correspondence: [email protected] The prosurvival transcription factor NF-kB specifically binds promoter DNA to activate target gene expression. NF-kB is regulated through interactions with IkB inhibitor proteins. Active proteolysis of these IkB proteins is, in turn, under the control of the IkB kinase complex (IKK). Together, these three molecules form the NF-kB signaling module. Studies aimed at charac- terizing the molecular mechanisms of NF-kB, IkB, and IKK in terms of their three-dimen- sional structures have lead to a greater understanding of this vital transcription factor system. F-kB is a master transcription factor that from the perspective of their three-dimensional Nresponds to diverse cell stimuli by activat- structures. ing the expression of stress response genes. Multiple signals, including cytokines, growth factors, engagement of the T-cell receptor, and NF-kB bacterial and viral products, induce NF-kB Introduction to NF-kB transcriptional activity (Hayden and Ghosh 2008). A point of convergence for the myriad NF-kB was discovered in the laboratory of of NF-kB inducing signals is the IkB kinase David Baltimore as a nuclear activity with bind- complex (IKK). Active IKK in turn controls ing specificity toward a ten-base-pair DNA transcription factor NF-kB by regulating pro- sequence 50-GGGACTTTCC-30 present within teolysis of the IkB inhibitor protein (Fig. -
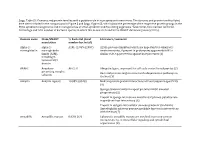
Supp. Table S2: Domains and Protein Families with a Putative Role in Host-Symbiont Interactions
Supp. Table S2: Domains and protein families with a putative role in host-symbiont interactions. The domains and protein families listed here were included in the comparisons in Figure 5 and Supp. Figure S5, which show the percentage of the respective protein groups in the Riftia symbiont metagenome and in metagenomes of other symbiotic and free-living organisms. % bacterial, total number bacterial: Percentage and total number of bacterial species in which this domain is found in the SMART database (January 2019). Domain name Pfam/SMART % bacterial (total Literature/comment annotation number bacterial) Alpha-2- alpha-2- A2M: 42.05% (2057) A2Ms: protease inhibitors which are important for eukaryotic macroglobulin macroglobulin innate immunity, if present in prokaryotes apparently fulfill a family (A2M), similar role, e.g. protection against host proteases (1) including N- terminal MG1 domain ANAPC Anaphase- APC2: 0 Ubiquitin ligase, important for cell cycle control in eukaryotes (2) promoting complex Bacterial proteins might interact with ubiquitination pathways in subunits the host (3) Ankyrin Ankyrin repeats 10.88% (8348) Mediate protein-protein interactions without sequence specificity (4) Sponge symbiont ankyrin-repeat proteins inhibit amoebal phagocytosis (5) Present in sponge microbiome metatranscriptomes, putative role in symbiont-host interactions (6) Present in obligate intracellular amoeba symbiont Candidatus Amoebophilus asiaticus genome, probable function in interactions with the host (7) Armadillo Armadillo repeats 0.83% (67)