Liver Transcriptomics Highlights Interleukin-32 As Novel NAFLD
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Early Detection of Peripheral Blood Cell Signature in Children Developing B-Cell Autoimmunity at a Young Age
2024 Diabetes Volume 68, October 2019 Early Detection of Peripheral Blood Cell Signature in Children Developing b-Cell Autoimmunity at a Young Age Henna Kallionpää,1 Juhi Somani,2 Soile Tuomela,1 Ubaid Ullah,1 Rafael de Albuquerque,1 Tapio Lönnberg,1 Elina Komsi,1 Heli Siljander,3,4 Jarno Honkanen,3,4 Taina Härkönen,3,4 Aleksandr Peet,5,6 Vallo Tillmann,5,6 Vikash Chandra,3,7 Mahesh Kumar Anagandula,8 Gun Frisk,8 Timo Otonkoski,3,7 Omid Rasool,1 Riikka Lund,1 Harri Lähdesmäki,2 Mikael Knip,3,4,9,10 and Riitta Lahesmaa1 Diabetes 2019;68:2024–2034 | https://doi.org/10.2337/db19-0287 The appearance of type 1 diabetes (T1D)-associated function before T1D and suggest a potential role for IL32 autoantibodies is the first and only measurable param- in the pathogenesis of T1D. eter to predict progression toward T1D in genetically susceptible individuals. However, autoantibodies indi- cate an active autoimmune reaction, wherein the im- Family and sibling studies in type 1 diabetes (T1D) have mune tolerance is already broken. Therefore, there is implicated a firm genetic predisposition to a locus con- a clear and urgent need for new biomarkers that predict taining HLA class I and class II genes on chromosome the onset of the autoimmune reaction preceding auto- 6 suggesting a role for CD4+ as well as CD8+ T cells in T1D fl antibody positivity or re ect progressive b-cell destruc- pathogenesis (1–3). As much as 30–50% of the genetic risk – tion. Here we report the mRNA sequencing based is conferred by HLA class II molecules, which are crucial in analysis of 306 samples including fractionated samples antigen presentation to CD4+ T cells. -

IL-32 and IL-17 Interact and Have the Potential to Aggravate
Moon et al. Arthritis Research & Therapy 2012, 14:R246 http://arthritis-research.com/content/14/6/R246 RESEARCH ARTICLE Open Access IL-32 and IL-17 interact and have the potential to aggravate osteoclastogenesis in rheumatoid arthritis Young-Mee Moon1†, Bo-Young Yoon2†, Yang-Mi Her1, Hye-Joa Oh1, Jae-Seon Lee1, Kyoung-Woon Kim3, Seon-Yeong Lee1, Yun-Ju Woo1, Kyung-Su Park1, Sung-Hwan Park1, Ho-Youn Kim1 and Mi-La Cho1* Abstract Introduction: Interleukin (IL)-32 and IL-17 play critical roles in pro-inflammatory responses and are highly expressed in the synovium of patients with rheumatoid arthritis (RA). We investigated the relations between these two cytokines (IL-17 and IL-32) for their ability to induce each other and to stimulate osteoclasts in RA fibroblast- like synoviocytes (FLSs) and T cells. Methods: FLSs were isolated through surgical synovectomy obtained from patients with RA or osteoarthritis (OA). Real-time PCR were performed to evaluate the expression of IL-32, IL-17 and osteoclast-related genes. Immunohistochemical staining and tartrate-resistant acid phosphatase (TRAP) staining were performed to determine the distribution of inflammatory cytokines and the presence of osteoclastogenesis. Results: IL-17 induced the expression of IL-32 in the FLSs from RA patients, as assessed by microarray. IL-32 production was increased by IL-17. IL-32 in the FLSs from RA patients induced the production of IL-17 in CD4+ T cells. IL-32 and IL-17 were colocalized near TRAP-positive areas in joint specimens. IL-17 and IL-32 synergistically induced the differentiation of osteoclasts, as demonstrated by the expression of osteoclast-related genes. -

Evolutionary Divergence and Functions of the Human Interleukin (IL) Gene Family Chad Brocker,1 David Thompson,2 Akiko Matsumoto,1 Daniel W
UPDATE ON GENE COMPLETIONS AND ANNOTATIONS Evolutionary divergence and functions of the human interleukin (IL) gene family Chad Brocker,1 David Thompson,2 Akiko Matsumoto,1 Daniel W. Nebert3* and Vasilis Vasiliou1 1Molecular Toxicology and Environmental Health Sciences Program, Department of Pharmaceutical Sciences, University of Colorado Denver, Aurora, CO 80045, USA 2Department of Clinical Pharmacy, University of Colorado Denver, Aurora, CO 80045, USA 3Department of Environmental Health and Center for Environmental Genetics (CEG), University of Cincinnati Medical Center, Cincinnati, OH 45267–0056, USA *Correspondence to: Tel: þ1 513 821 4664; Fax: þ1 513 558 0925; E-mail: [email protected]; [email protected] Date received (in revised form): 22nd September 2010 Abstract Cytokines play a very important role in nearly all aspects of inflammation and immunity. The term ‘interleukin’ (IL) has been used to describe a group of cytokines with complex immunomodulatory functions — including cell proliferation, maturation, migration and adhesion. These cytokines also play an important role in immune cell differentiation and activation. Determining the exact function of a particular cytokine is complicated by the influence of the producing cell type, the responding cell type and the phase of the immune response. ILs can also have pro- and anti-inflammatory effects, further complicating their characterisation. These molecules are under constant pressure to evolve due to continual competition between the host’s immune system and infecting organisms; as such, ILs have undergone significant evolution. This has resulted in little amino acid conservation between orthologous proteins, which further complicates the gene family organisation. Within the literature there are a number of overlapping nomenclature and classification systems derived from biological function, receptor-binding properties and originating cell type. -
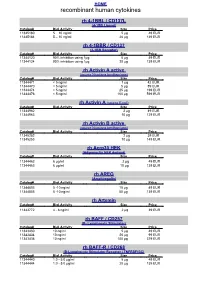
Recombinant Human Cytokines
HOME recombinant human cytokines rh 4-1BBL / CD137L (4-1BB Ligand) Catalog# Biol.Activity Size Price 11345180 5 – 10 ng/ml 5 µg 49 EUR 11345184 5 – 10 ng/ml 20 µg 139 EUR rh 4-1BBR / CD137 (4-1BB Receptor) Catalog# Biol.Activity Size Price 11344120 90% inhibition using 1µg 5 µg 49 EUR 11344124 90% inhibition using 1µg 20 µg 139 EUR rh Activin A active (source Nicotiana benthamiana) Catalog# Biol.Activity Size Price 11344471 < 5 ng/ml 1 µg 42 EUR 11344470 < 5 ng/ml 5 µg 89 EUR 11344474 < 5 ng/ml 25 µg 199 EUR 11344476 < 5 ng/ml 100 µg 599 EUR rh Activin A (source E.coli) Catalog# Biol.Activity Size Price 11344962 2 µg 49 EUR 11344963 10 µg 129 EUR rh Activin B active (source Nicotiana benthamiana) Catalog# Biol.Activity Size Price 11345252 2 µg 39 EUR 11345253 10 µg 149 EUR rh Acrp30 HEK (Adiponectin HEK derived) Catalog# Biol.Activity Size Price 11344462 6 µg/ml 2 µg 49 EUR 11344463 6 µg/ml 10 µg 139 EUR rh AREG (Amphiregulin) Catalog# Biol.Activity Size Price 11344803 5 -10 ng/ml 10 µg 49 EUR 11344805 5 -10 ng/ml 50 µg 139 EUR rh Artemin Catalog# Biol.Activity Size Price 11343772 4 - 8 ng/ml 2 µg 39 EUR rh BAFF / CD257 (B- Lymphocyte Stimulator) Catalog# Biol.Activity Size Price 11343430 10 ng/ml 5 µg 49 EUR 11343434 10 ng/ml 20 µg 99 EUR 11343436 10 ng/ml 100 µg 379 EUR rh BAFF-R / CD268 (B-Lymphocyte Stimulator Receptor / TNFRSF13C) Catalog# Biol.Activity Size Price 11344440 1.0 - 5.0 µg/ml 5 µg 49 EUR 11344444 1.0 - 5.0 µg/ml 20 µg 139 EUR HOME recombinant human cytokines rh BCA-1 (CXCL13) Catalog# Biol.Activity Size Price 11344180 -

Inflammatory Modulation of Hematopoietic Stem Cells by Magnetic Resonance Imaging
Electronic Supplementary Material (ESI) for RSC Advances. This journal is © The Royal Society of Chemistry 2014 Inflammatory modulation of hematopoietic stem cells by Magnetic Resonance Imaging (MRI)-detectable nanoparticles Sezin Aday1,2*, Jose Paiva1,2*, Susana Sousa2, Renata S.M. Gomes3, Susana Pedreiro4, Po-Wah So5, Carolyn Ann Carr6, Lowri Cochlin7, Ana Catarina Gomes2, Artur Paiva4, Lino Ferreira1,2 1CNC-Center for Neurosciences and Cell Biology, University of Coimbra, Coimbra, Portugal, 2Biocant, Biotechnology Innovation Center, Cantanhede, Portugal, 3King’s BHF Centre of Excellence, Cardiovascular Proteomics, King’s College London, London, UK, 4Centro de Histocompatibilidade do Centro, Coimbra, Portugal, 5Department of Neuroimaging, Institute of Psychiatry, King's College London, London, UK, 6Cardiac Metabolism Research Group, Department of Physiology, Anatomy & Genetics, University of Oxford, UK, 7PulseTeq Limited, Chobham, Surrey, UK. *These authors contributed equally to this work. #Correspondence to Lino Ferreira ([email protected]). Experimental Section Preparation and characterization of NP210-PFCE. PLGA (Resomers 502 H; 50:50 lactic acid: glycolic acid) (Boehringer Ingelheim) was covalently conjugated to fluoresceinamine (Sigma- Aldrich) according to a protocol reported elsewhere1. NPs were prepared by dissolving PLGA (100 mg) in a solution of propylene carbonate (5 mL, Sigma). PLGA solution was mixed with perfluoro- 15-crown-5-ether (PFCE) (178 mg) (Fluorochem, UK) dissolved in trifluoroethanol (1 mL, Sigma). This solution was then added to a PVA solution (10 mL, 1% w/v in water) dropwise and stirred for 3 h. The NPs were then transferred to a dialysis membrane and dialysed (MWCO of 50 kDa, Spectrum Labs) against distilled water before freeze-drying. Then, NPs were coated with protamine sulfate (PS). -

Interleukin-32 in Systemic Sclerosis, a Potential New Biomarker for Pulmonary Arterial Hypertension
Di Benedetto et al. Arthritis Research & Therapy (2020) 22:127 https://doi.org/10.1186/s13075-020-02218-8 RESEARCH ARTICLE Open Access Interleukin-32 in systemic sclerosis, a potential new biomarker for pulmonary arterial hypertension Paola Di Benedetto1†, Giuliana Guggino2†, Giovanna Manzi3, Piero Ruscitti4, Onorina Berardicurti4, Noemi Panzera1, Nicolò Grazia1, Roberto Badagliacca3, Valeria Riccieri5, Carmine Dario Vizza3, Ganna Radchenko6, Vasiliki Liakouli7, Francesco Ciccia7, Paola Cipriani4† and Roberto Giacomelli4*† Abstract Background: Pulmonary arterial hypertension (PAH) is a severe complication of systemic sclerosis (SSc), associated with a progressive elevation in pulmonary vascular resistance and subsequent right heart failure and death. Due to unspecific symptoms, the diagnosis of PAH is often delayed. On this basis, it is of great value to improve current diagnostic methods and develop new strategies for evaluating patients with suspected PAH. Interleukin-32 (IL-32) is a proinflammatory cytokine expressed in damaged vascular cells, and the present study aimed to assess if this cytokine could be a new biomarker of PAH during SSc. Methods: The IL-32 expression was evaluated in the sera and skin samples of 18 SSc-PAH patients, 21 SSc patients without PAH, 15 patients with idiopathic PAH (iPAH) and 14 healthy controls (HCs), by enzyme-linked immunosorbent assay (ELISA) and immunohistochemistry (IHC). Receiver-operating characteristic (ROC) curves were performed to evaluate the cut-off of IL-32 in identifying patients with PAH. Furthermore, in SSc patients, correlation analyses were performed between IL-32 sera levels and mean pulmonary artery pressure (mPAP) evaluated by right heart catheterization (RHC) and systolic pulmonary artery pressure (sPAP), obtained by echocardiography. -

Interleukin-32 Increases Human Gastric Cancer Cell Invasion Associated with Tumor Progression and Metastasis
Published OnlineFirst March 6, 2014; DOI: 10.1158/1078-0432.CCR-13-1221 Clinical Cancer Human Cancer Biology Research Interleukin-32 Increases Human Gastric Cancer Cell Invasion Associated with Tumor Progression and Metastasis Chung-Ying Tsai1, Chia-Siu Wang4, Ming-Ming Tsai2, Hsiang-Cheng Chi1, Wan-Li Cheng3, Yi-Hsin Tseng1, Cheng-Yi Chen1, Crystal D. Lin6, Jun-I. Wu5, Lu-Hai Wang5, and Kwang-Huei Lin1 Abstract Purpose: The proinflammatory cytokine interleukin-32 (IL-32) is a novel tumor marker highly expressed in various human carcinomas, including gastric cancer. However, its effects on prognosis of patients with gastric cancer and cancer metastasis are virtually unknown at present. The main aim of this study was to explore the clinical significance of IL-32 in gastric cancer and further elucidate the molecular mechanisms underlying IL-32–mediated migration and invasion. Experimental Design: Gastric cancer cells with ectopic expression or silencing of IL-32 were examined to identify downstream molecules and establish their effects on cell motility, invasion, and lung metastasis in vivo. Results: IL-32 was significantly upregulated in gastric cancer and positively correlated with aggressiveness of cancer and poor prognosis. Ectopic expression of IL-32 induced elongated morphology and increased cell migration and invasion via induction of IL-8, VEGF, matrix metalloproteinase 2 (MMP2), and MMP9 expression via phosphor-AKT/phospho-glycogen synthase kinase 3b/active b-catenin as well as hypoxia- inducible factor 1a (HIF-1a) signaling pathways. Conversely, depletion of IL-32 in gastric cancer cells reversed these effects and decreased lung colonization in vivo. Examination of gene expression datasets in oncomine and staining of gastric cancer specimens demonstrated the clinical significance of IL-32 and its downstream molecules by providing information on their coexpression patterns. -

Association Between Interleukin-32 and Interleukin-17A Single Nucleotide Polymorphisms and Serum Levels with Polycystic Ovary Syndrome
ORIGINAL ARTICLE Iran J Allergy Asthma Immunol February 2019; 18(1):91-99. Association between Interleukin-32 and Interleukin-17A Single Nucleotide Polymorphisms and Serum Levels with Polycystic Ovary Syndrome Fatemeh Hesampour1, Bahia Namavar Jahromi2,3, Foroozan Tahmasebi1, and Behrouz Gharesi-Fard1,2 1 Department of Immunology, Shiraz University of Medical Sciences, Shiraz, Iran 2 Infertility Research Center, Shiraz University of Medical Sciences, Shiraz, Iran 3 Department of Gynecology and Obstetrics, Shiraz University of Medical Sciences, Shiraz, Iran Received: 31 October 2018; Received in revised form: 3 December 2018; Accepted: 29 December 2018 ABSTRACT Polycystic ovary syndrome (PCOS) is correlated with low-grade chronic inflammation. Interleukin-17A (IL-17A) and Interleukin-32 (IL-32) are two members of the pro- inflammatory cytokines which act as significant components of the immune system during certain inflammatory diseases. Along with immunological processes, genetic factors play major roles in predisposition to PCOS. There are myriad single nucleotide polymorphisms (SNPs) within IL-17A and IL-32 genes that may affect their production and the susceptibility of individuals to PCOS. The objective of the present research was to investigate the association between IL-17A (rs2275913) and IL-32 (rs9927163, rs4786370) SNPs, and also their serum levels with susceptibility to PCOS in a group of Iranian women. In this case-control study, 150 PCOS patients (mean age of 29.1 years) and 150 healthy women (mean age of 26.1 years) were analyzed in terms of IL-17A and IL-32 SNPs via polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique. Furthermore, serum levels of IL-17A and IL-32 cytokines were measured through the use of ELISA method. -

IL-32 Protection from RNA and DNA Viruses By
Protection from RNA and DNA Viruses by IL-32 Jarod A. Zepp, Claudia A. Nold-Petry, Charles A. Dinarello and Marcel F. Nold This information is current as of October 1, 2021. J Immunol 2011; 186:4110-4118; Prepublished online 23 February 2011; doi: 10.4049/jimmunol.1000081 http://www.jimmunol.org/content/186/7/4110 Downloaded from References This article cites 42 articles, 15 of which you can access for free at: http://www.jimmunol.org/content/186/7/4110.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on October 1, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Protection from RNA and DNA Viruses by IL-32 Jarod A. Zepp,* Claudia A. Nold-Petry,*,† Charles A. Dinarello,* and Marcel F. Nold*,† Several studies have documented a proinflammatory role for IL-32, which induces IL-1a, IL-1b, IL-6, TNF, and chemokines via NF-kB, p38MAPK, and AP-1. -

Human Cytokine Response Profiles
Comprehensive Understanding of the Human Cytokine Response Profiles A. Background The current project aims to collect datasets profiling gene expression patterns of human cytokine treatment response from the NCBI GEO and EBI ArrayExpress databases. The Framework for Data Curation already hosted a list of candidate datasets. You will read the study design and sample annotations to select the relevant datasets and label the sample conditions to enable automatic analysis. If you want to build a new data collection project for your topic of interest instead of working on our existing cytokine project, please read section D. We will explain the cytokine project’s configurations to give you an example on creating your curation task. A.1. Cytokine Cytokines are a broad category of small proteins mediating cell signaling. Many cell types can release cytokines and receive cytokines from other producers through receptors on the cell surface. Despite some overlap in the literature terminology, we exclude chemokines, hormones, or growth factors, which are also essential cell signaling molecules. Meanwhile, we count two cytokines in the same family as the same if they share the same receptors. In this project, we will focus on the following families and use the member symbols as standard names (Table 1). Family Members (use these symbols as standard cytokine names) Colony-stimulating factor GCSF, GMCSF, MCSF Interferon IFNA, IFNB, IFNG Interleukin IL1, IL1RA, IL2, IL3, IL4, IL5, IL6, IL7, IL9, IL10, IL11, IL12, IL13, IL15, IL16, IL17, IL18, IL19, IL20, IL21, IL22, IL23, IL24, IL25, IL26, IL27, IL28, IL29, IL30, IL31, IL32, IL33, IL34, IL35, IL36, IL36RA, IL37, TSLP, LIF, OSM Tumor necrosis factor TNFA, LTA, LTB, CD40L, FASL, CD27L, CD30L, 41BBL, TRAIL, OPGL, APRIL, LIGHT, TWEAK, BAFF Unassigned TGFB, MIF Table 1. -

RT² Profiler PCR Array (384-Well Format) Human Inflammatory Response & Autoimmunity
RT² Profiler PCR Array (384-Well Format) Human Inflammatory Response & Autoimmunity Cat. no. 330231 PAHS-3803ZE For pathway expression analysis Format For use with the following real-time cyclers RT² Profiler PCR Array, Applied Biosystems® models 7900HT (384-well block), Format E ViiA™ 7 (384-well block); Bio-Rad CFX384™ RT² Profiler PCR Array, Roche® LightCycler® 480 (384-well block) Format G Description The Human Inflammatory Response & Autoimmunity RT² Profiler PCR Array profiles the expression of 370 key genes involved in immune response during autoimmunity and inflammation. It represents the expression of inflammatory cytokines, chemokines, and their receptors. It also contains genes related to the production and metabolism of cytokines. Genes involved in cytokine-cytokine receptor interactions are as well as thoroughly researched panels of genes involved in the acute-phase, inflammatory, and humoral immune responses. Using real-time PCR, you can easily and reliably analyze the expression of a focused panel of genes related to autoimmunity and inflammation with this array. For further details, consult the RT² Profiler PCR Array Handbook. Sample & Assay Technologies Shipping and storage RT² Profiler PCR Arrays in formats E and G are shipped at ambient temperature, on dry ice, or blue ice packs depending on destination and accompanying products. For long term storage, keep plates at –20°C. Note: Ensure that you have the correct RT² Profiler PCR Array format for your real-time cycler (see table above). Note: Open the package and store -

MOL ID MOL Name OB DL Herb Source 1 MOL011616 Fortunellin
Table S1. Detailed information of active compounds (OB ≥ 30% and DL ≥ 0.18) in PHF MOL ID MOL Name OB DL Herb source 1 MOL011616 Fortunellin 35.65 0.74 BH 2 MOL001689 acacetin 34.97 0.24 BH 3 MOL001790 Linarin 39.84 0.71 BH 4 MOL002881 Diosmetin 31.14 0.27 BH 5 MOL000359 sitosterol 36.91 0.75 BH、DP 6 MOL004328 naringenin 59.29 0.21 BH 7 MOL000471 aloe-emodin 83.38 0.24 BH 8 MOL005190 eriodictyol 71.79 0.24 BH 9 MOL005573 Genkwanin 37.13 0.24 BH 10 MOL000006 luteolin 36.16 0.25 BH、JYH 11 MOL000173 wogonin 30.68 0.23 CZ 2-Hydroxyisoxypropyl-3-hydroxy-7- 12 MOL000179 isopentene-2,3-dihydrobenzofuran-5- 45.2 0.2 CZ carboxylic 13 MOL000184 NSC63551 39.25 0.76 CZ Stigmasterol 3-O-beta-D- 14 MOL000186 43.83 0.76 CZ glucopyranoside_qt 15 MOL000188 3β-acetoxyatractylone 40.57 0.22 CZ 16 MOL000085 beta-daucosterol_qt 36.91 0.75 CZ 17 MOL000088 beta-sitosterol 3-O-glucoside_qt 36.91 0.75 CZ 18 MOL000092 daucosterin_qt 36.91 0.76 CZ 19 MOL000094 daucosterol_qt 36.91 0.76 CZ 20 MOL001925 paeoniflorin_qt 68.18 0.4 DP 21 MOL000211 Mairin 55.38 0.78 DP 22 MOL000422 kaempferol 41.88 0.24 DP、JYH 23 MOL000492 (+)-catechin 54.83 0.24 DP 24 MOL007003 benzoyl paeoniflorin 31.14 0.54 DP 25 MOL007369 4-O-methylpaeoniflorin_qt 67.24 0.43 DP 5-[[5-(4-methoxyphenyl)-2- 26 MOL007374 43.44 0.3 DP furyl]methylene]barbituric acid 27 MOL007382 mudanpioside-h_qt 2 42.36 0.37 DP 28 MOL007384 paeonidanin_qt 65.31 0.35 DP 29 MOL000098 quercetin 46.43 0.28 DP、JYH、HB 30 MOL001454 berberine 36.86 0.78 HB 31 MOL001458 coptisine 30.67 0.86 HB 32 MOL002636 Kihadalactone A 34.21