Sea Shell Diversity and Rapidly Evolving Secretomes: Insights Into the Evolution of Biomineralization Kevin M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Can You Engineer an Insect Exoskeleton?
Page 1 of 14 Can you Engineer an Insect Exoskeleton? Submitted by Catherine Dana and Christina Silliman EnLiST Entomology Curriculum Developers Department of Entomology, University of Illinois Grade level targeted: 4th Grade, but can be easily adapted for later grades Big ideas: Insect exoskeleton, engineering, and biomimicry Main objective: Students will be able to design a functional model of an insect exoskeleton which meets specific physical requirements based on exoskeleton biomechanics Lesson Summary We humans have skin and bones to protect us and to help us stand upright. Insects don’t have bones or skin but they are protected from germs, physical harm, and can hold their body up on their six legs. This is all because of their hard outer shell, also known as their exoskeleton. This hard layer does more than just protect insects from being squished, and students will get to explore some of the many ways the exoskeleton protects insects by building one themselves! For this fourth grade lesson, students will work together in teams to use what they learn about exoskeleton biomechanics to design and build a protective casing. To complete the engineering design cycle, students can use what they learn from testing their case to redesign and re-build their prototype. Prerequisites No prior knowledge is required for this lesson. Students should be introduced to the general form of an insect to facilitate identification of the exoskeleton. Live insects would work best for this, but pictures and diagrams will work as well. Instruction Time 45 – 60 minutes Next Generation Science Standards (NGSS) Framework Alignment Disciplinary Core Ideas LS1.A: Structure and Function o Plants and animals have both internal and external structures that serve various functions in growth, survival, behavior, and reproduction. -
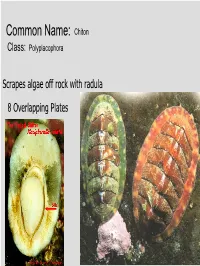
Common Name: Chiton Class: Polyplacophora
Common Name: Chiton Class: Polyplacophora Scrapes algae off rock with radula 8 Overlapping Plates Phylum? Mollusca Class? Gastropoda Common name? Brown sea hare Class? Scaphopoda Common name? Tooth shell or tusk shell Mud Tentacle Foot Class? Gastropoda Common name? Limpet Phylum? Mollusca Class? Bivalvia Class? Gastropoda Common name? Brown sea hare Phylum? Mollusca Class? Gastropoda Common name? Nudibranch Class? Cephalopoda Cuttlefish Octopus Squid Nautilus Phylum? Mollusca Class? Gastropoda Most Bivalves are Filter Feeders A B E D C • A: Mantle • B: Gill • C: Mantle • D: Foot • E: Posterior adductor muscle I.D. Green: Foot I.D. Red Gills Three Body Regions 1. Head – Foot 2. Visceral Mass 3. Mantle A B C D • A: Radula • B: Mantle • C: Mouth • D: Foot What are these? Snail Radulas Dorsal HingeA Growth line UmboB (Anterior) Ventral ByssalC threads Mussel – View of Outer Shell • A: Hinge • B: Umbo • C: Byssal threads Internal Anatomy of the Bay Mussel A B C D • A: Labial palps • B: Mantle • C: Foot • D: Byssal threads NacreousB layer Posterior adductorC PeriostracumA muscle SiphonD Mantle Byssal threads E Internal Anatomy of the Bay Mussel • A: Periostracum • B: Nacreous layer • C: Posterior adductor muscle • D: Siphon • E: Mantle Byssal gland Mantle Gill Foot Labial palp Mantle Byssal threads Gill Byssal gland Mantle Foot Incurrent siphon Byssal Labial palp threads C D B A E • A: Foot • B: Gills • C: Posterior adductor muscle • D: Excurrent siphon • E: Incurrent siphon Heart G F H E D A B C • A: Foot • B: Gills • C: Mantle • D: Excurrent siphon • E: Incurrent siphon • F: Posterior adductor muscle • G: Labial palps • H: Anterior adductor muscle Siphon or 1. -

Do You Think Animals Have Skeletons Like Ours?
Animal Skeletons Do you think animals have skeletons like ours? Are there any bones which might be similar? Vertebrate or Invertebrate § Look at the words above… § What do you think the difference is? § Hint: Break the words up (Vertebrae) Vertebrates and Invertebrates The difference between vertebrates and invertebrates is simple! Vertebrates have a backbone (spine)… …and invertebrates don’t Backbone (spine) vertebrate invertebrate So, if the animal has a backbone or a ‘vertebral column’ it is a ‘Vertebrate’ and if it doesn’t, it is called an ‘Invertebrate.’ It’s Quiz Time!! Put this PowerPoint onto full slideshow before starting. You will be shown a series of animals, click if you think it is a ‘Vertebrate’ or an ‘Invertebrate.’ Dog VertebrateVertebrate or InvertebrateInvertebrate Worm VertebrateVertebrate or InvertebrateInvertebrate Dinosaur VertebrateVertebrate or InvertebrateInvertebrate Human VertebrateVertebrate or InvertebrateInvertebrate Fish VertebrateVertebrate or InvertebrateInvertebrate Jellyfish VertebrateVertebrate or InvertebrateInvertebrate Butterfly VertebrateVertebrate or InvertebrateInvertebrate Types of Skeleton § Now we know the difference between ‘Vertebrate’ and ‘Invertebrate.’ § Let’s dive a little deeper… A further classification of skeletons comes from if an animal has a skeleton and where it is. All vertebrates have an endoskeleton. However invertebrates can be divided again between those with an exoskeleton and those with a hydrostatic skeleton. vertebrate invertebrate endoskeleton exoskeleton hydrostatic skeleton What do you think the words endoskeleton, exoskeleton and hydrostatic skeleton mean? Endoskeletons Animals with endoskeletons have Endoskeletons are lighter skeletons on the inside than exoskeletons. of their bodies. As the animal grows so does their skeleton. Exoskeletons Animals with exoskeletons Watch the following have clip to see how they shed their skeletons on their skeletons the outside! (clip the crab below). -

Marine Bivalve Molluscs
Marine Bivalve Molluscs Marine Bivalve Molluscs Second Edition Elizabeth Gosling This edition first published 2015 © 2015 by John Wiley & Sons, Ltd First edition published 2003 © Fishing News Books, a division of Blackwell Publishing Registered Office John Wiley & Sons, Ltd, The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK Editorial Offices 9600 Garsington Road, Oxford, OX4 2DQ, UK The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, UK 111 River Street, Hoboken, NJ 07030‐5774, USA For details of our global editorial offices, for customer services and for information about how to apply for permission to reuse the copyright material in this book please see our website at www.wiley.com/wiley‐blackwell. The right of the author to be identified as the author of this work has been asserted in accordance with the UK Copyright, Designs and Patents Act 1988. All rights reserved. No part of this publication may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, except as permitted by the UK Copyright, Designs and Patents Act 1988, without the prior permission of the publisher. Designations used by companies to distinguish their products are often claimed as trademarks. All brand names and product names used in this book are trade names, service marks, trademarks or registered trademarks of their respective owners. The publisher is not associated with any product or vendor mentioned in this book. Limit of Liability/Disclaimer of Warranty: While the publisher and author(s) have used their best efforts in preparing this book, they make no representations or warranties with respect to the accuracy or completeness of the contents of this book and specifically disclaim any implied warranties of merchantability or fitness for a particular purpose. -

Four Hundred Million Years of Silica Biomineralization in Land Plants
Four hundred million years of silica biomineralization in land plants Elizabeth Trembath-Reicherta,1, Jonathan Paul Wilsonb, Shawn E. McGlynna,c, and Woodward W. Fischera aDivision of Geological and Planetary Sciences, California Institute of Technology, Pasadena, CA 91125; bDepartment of Biology, Haverford College, Haverford, PA 19041; and cGraduate School of Science and Engineering, Tokyo Metropolitan University, Hachioji-shi, Tokyo 192-0397, Japan Edited by Thure E. Cerling, University of Utah, Salt Lake City, UT, and approved February 20, 2015 (received for review January 7, 2015) Biomineralization plays a fundamental role in the global silicon Silica is widely used within plants for structural support and cycle. Grasses are known to mobilize significant quantities of Si in pathogen defense (19–21), but it remains a poorly understood the form of silica biominerals and dominate the terrestrial realm aspect of plant biology. Recent work on the angiosperm Oryza today, but they have relatively recent origins and only rose to sativa demonstrated that silica accumulation is facilitated by taxonomic and ecological prominence within the Cenozoic Era. transmembrane proteins expressed in root cells (21–24). Phy- This raises questions regarding when and how the biological silica logenetic analysis revealed that these silicon transport proteins cycle evolved. To address these questions, we examined silica were derived from a diverse family of modified aquaporins that abundances of extant members of early-diverging land plant include arsenite and glycerol transporters (19, 21, 25, 26). A clades, which show that silica biomineralization is widespread different member of this aquaporin family was recently identi- across terrestrial plant linages. Particularly high silica abundances fied that enables silica uptake in the horsetail Equisetum,an are observed in lycophytes and early-diverging ferns. -

Haliotis Asinina) in Coastal Waters of Thailand Determined Using Microsatellite Markers
Mar. Biotechnol. 6, 604–611, 2004 DOI: 10.1007/s10126-004-2300-5 Ó 2005 Springer Science+Business Media, Inc. Population Structure of Tropical Abalone (Haliotis asinina) in Coastal Waters of Thailand Determined Using Microsatellite Markers S. Tang,1 A. Tassanakajon,1 S. Klinbunga,2 P. Jarayabhand,3,4 and P. Menasveta2,4 1Department of Biochemistry, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand 2Marine Biotechnology Research Unit, National Center for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency, Pathumthani 12120, Thailand 3Aquatic Resources Research Institute, Chulalongkorn University, Bangkok 10330, Thailand 4Department of Marine Science, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand Abstract: Three partial genomic libraries were constructed from genomic DNA of the tropical abalone (Haliotis asinina) that was digested with AluI, vortexed/sonicated, and digested with mixed enzyme (AluI, HincII, and RsaI). The libraries yielded 0.02%, 0.42%, and 1.46% positive microsatellite-containing clones, respectively. Eleven clones each of perfect, imperfect, and compound microsatellites were isolated. Ten primer pairs (CU- Has1–CUHas10) were analyzed to evaluate their polymorphic level. The numbers of alleles per locus, observed heterozygosity (H0), and expected heterozygosity (He) ranged from 3 to 26 alleles, and varied between 0.27 and 0.85 and between 0.24 and 0.93, respectively. Three microsatellite loci (CUHas2, CUHas3, and CUHas8) were further used for examination of genetic diversity and differentiation of natural H. asinina in coastal waters of Thailand. Genetic variabilities in terms of the effective number of alleles (ne), H0, and He were higher in 2 samples from the Gulf of Thailand (ne = 9.37, 7.66; H0 = 0.62, 0.78; and He = 0.87, 0.86) than those of one sample (ne = 6.04; H0 = 0.58; and He = 0.62) derived from the Andaman Sea. -

Clam Dissection Guideline
Clam Dissection Guideline BACKGROUND: Clams are bivalves, meaning that they have shells consisting of two halves, or valves. The valves are joined at the top, and the adductor muscles on each side hold the shell closed. If the adductor muscles are relaxed, the shell is pulled open by ligaments located on each side of the umbo. The clam's foot is used to dig down into the sand, and a pair of long incurrent and excurrent siphons that extrude from the clam's mantle out the side of the shell reach up to the water above (only the exit points for the siphons are shown). Clams are filter feeders. Water and food particles are drawn in through one siphon to the gills where tiny, hair-like cilia move the water, and the food is caught in mucus on the gills. From there, the food-mucus mixture is transported along a groove to the palps (mouth flaps) which push it into the clam's mouth. The second siphon carries away the water. The gills also draw oxygen from the water flow. The mantle, a thin membrane surrounding the body of the clam, secretes the shell. The oldest part of the clam shell is the umbo, and it is from the hinge area that the clam extends as it grows. I. Purpose: The purpose of this lab is to identify the internal and external structures of a mollusk by dissecting a clam. II. Materials: 2 pairs of safety goggles 1 paper towel 2 pairs of gloves 1 pair of scissors 1 preserved clam 2 pairs of forceps 1 dissecting tray 2 probes III. -

Download Book (PDF)
M o Manual on IDENTIFICATION OF SCHEDULE MOLLUSCS From India RAMAKRISHN~~ AND A. DEY Zoological Survey of India, M-Block, New Alipore, Kolkota 700 053 Edited by the Director, Zoological Survey of India, Kolkata ZOOLOGICAL SURVEY OF INDIA KOLKATA CITATION Ramakrishna and Dey, A. 2003. Manual on the Identification of Schedule Molluscs from India: 1-40. (Published : Director, Zool. Surv. India, Kolkata) Published: February, 2003 ISBN: 81-85874-97-2 © Government of India, 2003 ALL RIGHTS RESERVED • No part of this publication may be reproduced, stored in a retrieval system or transmitted, in any from or by any means, electronic, mechanical, photocopying, recording or otherwise without the prior permission of the publisher. • -This book is sold subject to the condition that it shall not, by way of trade, be lent, resold hired out or otherwise disposed of without the publisher's consent, in any form of binding or cover other than that in which it is published. • The correct price of this publication is the price printed on this page. Any revised price indicated by a rubber stamp or by a sticker or by any other means is incorrect and should be unacceptable. PRICE India : Rs. 250.00 Foreign : $ (U.S.) 15, £ 10 Published at the Publication Division by the Director, Zoological Survey of India, 234/4, AJ.C. Bose Road, 2nd MSO Building (13th Floor), Nizam Palace, Kolkata -700020 and printed at Shiva Offset, Dehra Dun. Manual on IDENTIFICATION OF SCHEDULE MOLLUSCS From India 2003 1-40 CONTENTS INTRODUcrION .............................................................................................................................. 1 DEFINITION ............................................................................................................................ 2 DIVERSITY ................................................................................................................................ 2 HA.B I,.-s .. .. .. 3 VAWE ............................................................................................................................................ -

Phylum MOLLUSCA Chitons, Bivalves, Sea Snails, Sea Slugs, Octopus, Squid, Tusk Shell
Phylum MOLLUSCA Chitons, bivalves, sea snails, sea slugs, octopus, squid, tusk shell Bruce Marshall, Steve O’Shea with additional input for squid from Neil Bagley, Peter McMillan, Reyn Naylor, Darren Stevens, Di Tracey Phylum Aplacophora In New Zealand, these are worm-like molluscs found in sandy mud. There is no shell. The tiny MOLLUSCA solenogasters have bristle-like spicules over Chitons, bivalves, sea snails, sea almost the whole body, a groove on the underside of the body, and no gills. The more worm-like slugs, octopus, squid, tusk shells caudofoveates have a groove and fewer spicules but have gills. There are 10 species, 8 undescribed. The mollusca is the second most speciose animal Bivalvia phylum in the sea after Arthropoda. The phylum Clams, mussels, oysters, scallops, etc. The shell is name is taken from the Latin (molluscus, soft), in two halves (valves) connected by a ligament and referring to the soft bodies of these creatures, but hinge and anterior and posterior adductor muscles. most species have some kind of protective shell Gills are well-developed and there is no radula. and hence are called shellfish. Some, like sea There are 680 species, 231 undescribed. slugs, have no shell at all. Most molluscs also have a strap-like ribbon of minute teeth — the Scaphopoda radula — inside the mouth, but this characteristic Tusk shells. The body and head are reduced but Molluscan feature is lacking in clams (bivalves) and there is a foot that is used for burrowing in soft some deep-sea finned octopuses. A significant part sediments. The shell is open at both ends, with of the body is muscular, like the adductor muscles the narrow tip just above the sediment surface for and foot of clams and scallops, the head-foot of respiration. -

Evolution of Large Body Size in Abalones (Haliotis): Patterns and Implications
Paleobiology, 31(4), 2005, pp. 591±606 Evolution of large body size in abalones (Haliotis): patterns and implications James A. Estes, David R. Lindberg, and Charlie Wray Abstract.ÐKelps and other ¯eshy macroalgaeÐdominant reef-inhabiting organisms in cool seasÐ may have radiated extensively following late Cenozoic polar cooling, thus triggering a chain of evolutionary change in the trophic ecology of nearshore temperate ecosystems. We explore this hypothesis through an analysis of body size in the abalones (Gastropoda; Haliotidae), a widely distributed group in modern oceans that displays a broad range of body sizes and contains fossil representatives from the late Cretaceous (60±75 Ma). Geographic analysis of maximum shell length in living abalones showed that small-bodied species, while most common in the Tropics, have a cosmopolitan distribution, whereas large-bodied species occur exclusively in cold-water ecosys- tems dominated by kelps and other macroalgae. The phylogeography of body size evolution in extant abalones was assessed by constructing a molecular phylogeny in a mix of large and small species obtained from different regions of the world. This analysis demonstrates that small body size is the plesiomorphic state and largeness has likely arisen at least twice. Finally, we compiled data on shell length from the fossil record to determine how (slowly or suddenly) and when large body size arose in the abalones. These data indicate that large body size appears suddenly at the Miocene/Pliocene boundary. Our ®ndings support the view that ¯eshy-algal dominated ecosys- tems radiated rapidly in the coastal oceans with the onset of the most recent glacial age. -

Biomineralization and Global Biogeochemical Cycles Philippe Van Cappellen Faculty of Geosciences, Utrecht University P.O
1122 Biomineralization and Global Biogeochemical Cycles Philippe Van Cappellen Faculty of Geosciences, Utrecht University P.O. Box 80021 3508 TA Utrecht, The Netherlands INTRODUCTION Biological activity is a dominant force shaping the chemical structure and evolution of the earth surface environment. The presence of an oxygenated atmosphere- hydrosphere surrounding an otherwise highly reducing solid earth is the most striking consequence of the rise of life on earth. Biological evolution and the functioning of ecosystems, in turn, are to a large degree conditioned by geophysical and geological processes. Understanding the interactions between organisms and their abiotic environment, and the resulting coupled evolution of the biosphere and geosphere is a central theme of research in biogeology. Biogeochemists contribute to this understanding by studying the transformations and transport of chemical substrates and products of biological activity in the environment. Biogeochemical cycles provide a general framework in which geochemists organize their knowledge and interpret their data. The cycle of a given element or substance maps out the rates of transformation in, and transport fluxes between, adjoining environmental reservoirs. The temporal and spatial scales of interest dictate the selection of reservoirs and processes included in the cycle. Typically, the need for a detailed representation of biological process rates and ecosystem structure decreases as the spatial and temporal time scales considered increase. Much progress has been made in the development of global-scale models of biogeochemical cycles. Although these models are based on fairly simple representations of the biosphere and hydrosphere, they account for the large-scale changes in the composition, redox state and biological productivity of the earth surface environment that have occurred over geological time. -

Durham Research Online
Durham Research Online Deposited in DRO: 23 May 2017 Version of attached le: Accepted Version Peer-review status of attached le: Peer-reviewed Citation for published item: Betts, Marissa J. and Paterson, John R. and Jago, James B. and Jacquet, Sarah M. and Skovsted, Christian B. and Topper, Timothy P. and Brock, Glenn A. (2017) 'Global correlation of the early Cambrian of South Australia : shelly fauna of the Dailyatia odyssei Zone.', Gondwana research., 46 . pp. 240-279. Further information on publisher's website: https://doi.org/10.1016/j.gr.2017.02.007 Publisher's copyright statement: c 2017 This manuscript version is made available under the CC-BY-NC-ND 4.0 license http://creativecommons.org/licenses/by-nc-nd/4.0/ Additional information: Use policy The full-text may be used and/or reproduced, and given to third parties in any format or medium, without prior permission or charge, for personal research or study, educational, or not-for-prot purposes provided that: • a full bibliographic reference is made to the original source • a link is made to the metadata record in DRO • the full-text is not changed in any way The full-text must not be sold in any format or medium without the formal permission of the copyright holders. Please consult the full DRO policy for further details. Durham University Library, Stockton Road, Durham DH1 3LY, United Kingdom Tel : +44 (0)191 334 3042 | Fax : +44 (0)191 334 2971 https://dro.dur.ac.uk Accepted Manuscript Global correlation of the early Cambrian of South Australia: Shelly fauna of the Dailyatia odyssei Zone Marissa J.