Supplementary Table 7
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Note Hominoid Fission of Chromosome 14/15 and Role Of
Supplemental Note Supplemental Note Hominoid fission of chromosome 14/15 and role of segmental duplications Giuliana Giannuzzi, Michele Pazienza, John Huddleston, Francesca Antonacci, Maika Malig, Laura Vives, Evan E. Eichler and Mario Ventura 1. Analysis of the macaque contig spanning the hominoid 14/15 fission site We grouped contig clones based on their FISH pattern on human and macaque chromosomes (Groups F1, F2, F3, G1, G2, G3, and G4). Group F2 clones (Hsa15b- and Hsa15c-positive) showed a single signal on macaque 7q and three signal clusters on human chromosome 15: at the orthologous 15q26, as expected, as well as at 15q11–14 and 15q24–25, which correspond to the actual and ancestral pericentromeric regions, respectively (Ventura et al. 2003). Indeed, this locus contains an LCR15 copy (Pujana et al. 2001) in both the macaque and human genomes. Most BAC clones were one-end anchored in the human genome (chr15:100,028–100,071 kb). In macaque this region experienced a 64 kb duplicative insertion from chromosome 17 (orthologous to human chromosome 13) (Figure 2), with the human configuration (absence of insertion) likely being the ancestral state because it is identical in orangutan and marmoset. Four Hsa15b- positive clones mapped on macaque and human chromosome 19, but neither human nor macaque assemblies report the STS Hsa15b duplicated at this locus. The presence of assembly gaps in macaque may explain why the STS is not annotated in this region. We aligned the sequence of macaque CH250-70H12 (AC187495.2) versus its human orthologous sequence (hg18 chr15:100,039k-100,170k) and found a 12 kb human expansion through tandem duplication of a ~100 bp unit corresponding to a portion of exon 20 of DNM1 (Figure S3). -

Genetic Variation Across the Human Olfactory Receptor Repertoire Alters Odor Perception
bioRxiv preprint doi: https://doi.org/10.1101/212431; this version posted November 1, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Genetic variation across the human olfactory receptor repertoire alters odor perception Casey Trimmer1,*, Andreas Keller2, Nicolle R. Murphy1, Lindsey L. Snyder1, Jason R. Willer3, Maira Nagai4,5, Nicholas Katsanis3, Leslie B. Vosshall2,6,7, Hiroaki Matsunami4,8, and Joel D. Mainland1,9 1Monell Chemical Senses Center, Philadelphia, Pennsylvania, USA 2Laboratory of Neurogenetics and Behavior, The Rockefeller University, New York, New York, USA 3Center for Human Disease Modeling, Duke University Medical Center, Durham, North Carolina, USA 4Department of Molecular Genetics and Microbiology, Duke University Medical Center, Durham, North Carolina, USA 5Department of Biochemistry, University of Sao Paulo, Sao Paulo, Brazil 6Howard Hughes Medical Institute, New York, New York, USA 7Kavli Neural Systems Institute, New York, New York, USA 8Department of Neurobiology and Duke Institute for Brain Sciences, Duke University Medical Center, Durham, North Carolina, USA 9Department of Neuroscience, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA *[email protected] ABSTRACT The human olfactory receptor repertoire is characterized by an abundance of genetic variation that affects receptor response, but the perceptual effects of this variation are unclear. To address this issue, we sequenced the OR repertoire in 332 individuals and examined the relationship between genetic variation and 276 olfactory phenotypes, including the perceived intensity and pleasantness of 68 odorants at two concentrations, detection thresholds of three odorants, and general olfactory acuity. -

Database Tool the Systematic Annotation of the Three Main GPCR
Database, Vol. 2010, Article ID baq018, doi:10.1093/database/baq018 ............................................................................................................................................................................................................................................................................................. Database tool The systematic annotation of the three main Downloaded from https://academic.oup.com/database/article-abstract/doi/10.1093/database/baq018/406672 by guest on 15 January 2019 GPCR families in Reactome Bijay Jassal1, Steven Jupe1, Michael Caudy2, Ewan Birney1, Lincoln Stein2, Henning Hermjakob1 and Peter D’Eustachio3,* 1European Bioinformatics Institute, Hinxton, Cambridge, CB10 1SD, UK, 2Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada and 3New York University School of Medicine, New York, NY 10016, USA *Corresponding author: Tel: +212 263 5779; Fax: +212 263 8166; Email: [email protected] Submitted 14 April 2010; Revised 14 June 2010; Accepted 13 July 2010 ............................................................................................................................................................................................................................................................................................. Reactome is an open-source, freely available database of human biological pathways and processes. A major goal of our work is to provide an integrated view of cellular signalling processes that spans from ligand–receptor -

OR2AG1 (NM 001004489) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC214778 OR2AG1 (NM_001004489) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: OR2AG1 (NM_001004489) Human Tagged ORF Clone Tag: Myc-DDK Symbol: OR2AG1 Synonyms: OR2AG3; OR11-79 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC214778 representing NM_001004489 Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGGAGCTCTGGAACTTCACCTTGGGAAGTGGCTTCATTTTGGTGGGGATTCTGAATGACAGTGGGTCTC CTGAACTGCTCTGTGCTACAATTACAATCCTATACTTGTTGGCCCTGATCAGCAATGGCCTACTGCTCCT GGCTATCACCATGGAAGCCCGGCTCCACATGCCCATGTACCTCCTGCTTGGGCAGCTCTCTCTCATGGAC CTCCTGTTCACATCTGTTGTCACTCCCAAGGCCCTTGCGGACTTTCTGCGCAGAGAAAACACCATCTCCT TTGGAGGCTGTGCCCTTCAGATGTTCCTGGCACTGACAATGGGTGGTGCTGAGGACCTCCTACTGGCCTT CATGGCCTATGACAGGTATGTGGCCATTTGTCATCCTCTGACATACATGACCCTCATGAGCTCAAGAGCC TGCTGGCTCATGGTGGCCACGTCCTGGATCCTGGCATCCCTAAGTGCCCTAATATATACCGTGTATACCA TGCACTATCCCTTCTGCAGGGCCCAGGAGATCAGGCATCTTCTCTGTGAGATCCCACACTTGCTGAAGGT GGCCTGTGCTGATACCTCCAGATATGAGCTCATGGTATATGTGATGGGTGTGACCTTCCTGATTCCCTCT CTTGCTGCTATACTGGCCTCCTATACACAAATTCTACTCACTGTGCTCCATATGCCATCAAATGAGGGGA GGAAGAAAGCCCTTGTCACCTGCTCTTCCCACCTGACTGTGGTTGGGATGTTCTATGGAGCTGCCACATT CATGTATGTCTTGCCCAGTTCCTTCCACAGCACCAGACAAGACAACATCATCTCTGTTTTCTACACAATT GTCACTCCAGCCCTGAATCCACTCATCTACAGCCTGAGGAATAAGGAGGTCATGCGGGCCTTGAGGAGGG -

Assembly and Annotation of an Ashkenazi Human Reference Genome
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.18.997395; this version posted March 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Assembly and Annotation of an Ashkenazi Human Reference Genome Alaina Shumate1,2,† Aleksey V. Zimin1,2,† Rachel M. Sherman1,3 Daniela Puiu1,3 Justin M. Wagner4 Nathan D. Olson4 Mihaela Pertea1,2 Marc L. Salit5 Justin M. Zook4 Steven L. Salzberg1,2,3,6* 1Center for Computational Biology, Johns Hopkins University, Baltimore, MD 2Department of Biomedical Engineering, Johns Hopkins University, Baltimore, MD 3Department of Computer Science, Johns Hopkins University, Baltimore, MD 4National Institute of Standards and Technology, Gaithersburg, MD 5Joint Initiative for Metrology in Biology, Stanford University, Stanford, CA 6Department of Biostatistics, Johns Hopkins University, Baltimore, MD †These authors contributed equally to this work. *Corresponding author. Email: [email protected] Abstract Here we describe the assembly and annotation of the genome of an Ashkenazi individual and the creation of a new, population-specific human reference genome. This genome is more contiguous and more complete than GRCh38, the latest version of the human reference genome, and is annotated with highly similar gene content. The Ashkenazi reference genome, Ash1, contains 2,973,118,650 nucleotides as compared to 2,937,639,212 in GRCh38. Annotation identified 20,157 protein-coding genes, of which 19,563 are >99% identical to their counterparts on GRCh38. Most of the remaining genes have small differences. -

Genomic Selection Signatures in Sheep from the Western Pyrenees Otsanda Ruiz-Larrañaga, Jorge Langa, Fernando Rendo, Carmen Manzano, Mikel Iriondo, Andone Estonba
Genomic selection signatures in sheep from the Western Pyrenees Otsanda Ruiz-Larrañaga, Jorge Langa, Fernando Rendo, Carmen Manzano, Mikel Iriondo, Andone Estonba To cite this version: Otsanda Ruiz-Larrañaga, Jorge Langa, Fernando Rendo, Carmen Manzano, Mikel Iriondo, et al.. Genomic selection signatures in sheep from the Western Pyrenees. Genetics Selection Evolution, BioMed Central, 2018, 50 (1), pp.9. 10.1186/s12711-018-0378-x. hal-02405217 HAL Id: hal-02405217 https://hal.archives-ouvertes.fr/hal-02405217 Submitted on 11 Dec 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Ruiz-Larrañaga et al. Genet Sel Evol (2018) 50:9 https://doi.org/10.1186/s12711-018-0378-x Genetics Selection Evolution RESEARCH ARTICLE Open Access Genomic selection signatures in sheep from the Western Pyrenees Otsanda Ruiz‑Larrañaga1* , Jorge Langa1, Fernando Rendo2, Carmen Manzano1, Mikel Iriondo1 and Andone Estonba1 Abstract Background: The current large spectrum of sheep phenotypic diversity -

A Nonsense (C.3978G>A) Abnormal Spindle-Like, Microcephaly Associated (ASPM) Gene Mutation Is a Major Cause of Primary Microc
African Journal of Biotechnology Vol. 10(34), pp. 6396-6400, 11 July, 2011 Available online at http://www.academicjournals.org/AJB DOI: 10.5897/AJB10.2571 ISSN 1684-5315 © 2011 Academic Journals Full Length Research Paper A nonsense (c.3978G>A) abnormal spindle-like, microcephaly associated (ASPM) gene mutation is a major cause of primary microcephaly in Pashtoon ethnic group of Pakistan Shamim Saleha 1, Muhammad Ajmal 2, Muhammad Jamil 1, Muhammad Nasir 2 and Abdul Hameed 2* 1Department of Biotechnology and Genetic Engineering, Kohat University of Science and Technology, Kohat 26000, Khyber Paktoonkhwa, Pakistan. 2Institute of Biomedical and Genetic Engineering, G.P.O. box 2891, 24-Mauve Area, G-9/1, Islamabad, Pakistan. Accepted 29 April, 2011 Primary microcephaly (MCPH) is an autosomal-recessive congenital disorder characterized by smaller- than-normal brain size and mental retardation. MCPH is genetically heterogeneous with six known loci: MCPH1 to MCPH7. The abnormal spindle-like, microcephaly associated (ASPM) gene at MCPH5 locus, which accounts for 37 to 54% of MCPH, appears to be the most common cause of microcephaly. More than 50% of the MCPH families genetically analyzed in Pakistan were mapped to MCPH5 locus including both families in this study. On mutation screening of ASPM gene by PCR amplification and direct DNA sequencing, a common c.3978G>A transition was identified in exon 17 of ASPM gene to be responsible for diseased phenotype in both families. This change results to the substitution of an amino acid residue at position 1326 from tryptophan to a stop codon (p.Trp1326Stop). The same mutation was also identified in several other families of Pakistani origin. -

Molecular Genetics of Microcephaly Primary Hereditary: an Overview
brain sciences Review Molecular Genetics of Microcephaly Primary Hereditary: An Overview Nikistratos Siskos † , Electra Stylianopoulou †, Georgios Skavdis and Maria E. Grigoriou * Department of Molecular Biology & Genetics, Democritus University of Thrace, 68100 Alexandroupolis, Greece; [email protected] (N.S.); [email protected] (E.S.); [email protected] (G.S.) * Correspondence: [email protected] † Equal contribution. Abstract: MicroCephaly Primary Hereditary (MCPH) is a rare congenital neurodevelopmental disorder characterized by a significant reduction of the occipitofrontal head circumference and mild to moderate mental disability. Patients have small brains, though with overall normal architecture; therefore, studying MCPH can reveal not only the pathological mechanisms leading to this condition, but also the mechanisms operating during normal development. MCPH is genetically heterogeneous, with 27 genes listed so far in the Online Mendelian Inheritance in Man (OMIM) database. In this review, we discuss the role of MCPH proteins and delineate the molecular mechanisms and common pathways in which they participate. Keywords: microcephaly; MCPH; MCPH1–MCPH27; molecular genetics; cell cycle 1. Introduction Citation: Siskos, N.; Stylianopoulou, Microcephaly, from the Greek word µικρoκεϕαλi´α (mikrokephalia), meaning small E.; Skavdis, G.; Grigoriou, M.E. head, is a term used to describe a cranium with reduction of the occipitofrontal head circum- Molecular Genetics of Microcephaly ference equal, or more that teo standard deviations -
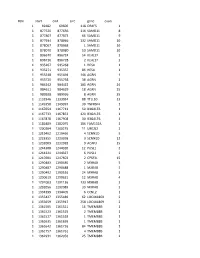
Chr Start End Size Gene Exon 1 69482 69600 118 OR4F5 1 1 877520
#chr start end size gene exon 1 69482 69600 118 OR4F5 1 1 877520 877636 116 SAMD11 8 1 877807 877873 66 SAMD11 9 1 877934 878066 132 SAMD11 10 1 878067 878068 1 SAMD11 10 1 878070 878080 10 SAMD11 10 1 896670 896724 54 KLHL17 2 1 896726 896728 2 KLHL17 2 1 935267 935268 1 HES4 1 1 935271 935357 86 HES4 1 1 955548 955694 146 AGRN 1 1 955720 955758 38 AGRN 1 1 984242 984422 180 AGRN 24 1 984611 984629 18 AGRN 25 1 989928 989936 8 AGRN 35 1 1132946 1133034 88 TTLL10 13 1 1149358 1149397 39 TNFRSF4 1 1 1167654 1167713 59 B3GALT6 1 1 1167733 1167853 120 B3GALT6 1 1 1167878 1167908 30 B3GALT6 1 1 1181889 1182075 186 FAM132A 1 1 1200204 1200215 11 UBE2J2 2 1 1219462 1219466 4 SCNN1D 5 1 1223355 1223358 3 SCNN1D 12 1 1232009 1232018 9 ACAP3 15 1 1244308 1244320 12 PUSL1 2 1 1244321 1244327 6 PUSL1 2 1 1247601 1247603 2 CPSF3L 15 1 1290483 1290485 2 MXRA8 5 1 1290487 1290488 1 MXRA8 5 1 1290492 1290516 24 MXRA8 5 1 1290619 1290631 12 MXRA8 4 1 1291003 1291136 133 MXRA8 3 1 1292056 1292089 33 MXRA8 2 1 1334399 1334405 6 CCNL2 1 1 1355427 1355489 62 LOC441869 2 1 1355659 1355917 258 LOC441869 2 1 1361505 1361521 16 TMEM88B 1 1 1361523 1361525 2 TMEM88B 1 1 1361527 1361528 1 TMEM88B 1 1 1361635 1361636 1 TMEM88B 1 1 1361642 1361726 84 TMEM88B 1 1 1361757 1361761 4 TMEM88B 1 1 1362931 1362956 25 TMEM88B 2 1 1374780 1374838 58 VWA1 3 1 1374841 1374842 1 VWA1 3 1 1374934 1375100 166 VWA1 3 1 1389738 1389747 9 ATAD3C 4 1 1389751 1389816 65 ATAD3C 4 1 1389830 1389849 19 ATAD3C 4 1 1390835 1390837 2 ATAD3C 5 1 1407260 1407331 71 ATAD3B 1 1 1407340 1407474 -

The Molecular Landscape of ASPM Mutations in Primary Microcephaly
Original article J Med Genet: first published as 10.1136/jmg.2008.062380 on 21 November 2008. Downloaded from The molecular landscape of ASPM mutations in primary microcephaly A K Nicholas,1 E A Swanson,2 J J Cox,1 G Karbani,3 S Malik,3 K Springell,4 D Hampshire,4 M Ahmed,3 J Bond,4 D Di Benedetto,5 M Fichera,5 C Romano,6 W B Dobyns,2 C G Woods1 c Additional tables are ABSTRACT also a diagnosable cause of mental retardation, and published online only at http:// Background: Autosomal recessive primary microcephaly one with a substantial recurrence risk of one in jmg.bmj.com/content/vol46/ (MCPH) is a model disease to study human neurogenesis. issue4 four in subsequent children. In affected individuals the brain grows at a reduced rate 1 The current diagnostic criteria for MCPH are: Department of Medical during fetal life resulting in a small but structurally normal congenital microcephaly more than 23 SD below Genetics, Cambridge Institute for Medical Research, University brain and mental retardation. The condition is genetically age and sex means; mental retardation but no of Cambridge, Cambridge, UK; heterogeneous with mutations in ASPM being most other neurological finding, such as spasticity, 2 University of Chicago, commonly reported. seizures, or progressive cognitive decline; normal Department of Human Genetics, Methods and results: We have examined this further by height and weight, appearance, and results on Chicago, Illinois, USA; studying three cohorts of microcephalic children to extend 6 3 Department of Clinical chromosome analysis and brain scan. Despite this, Genetics, St James’s University both the phenotype and the mutation spectrum. -

Genome-Wide Transcriptome Analysis of Laminar Tissue During the Early Stages of Experimentally Induced Equine Laminitis
GENOME-WIDE TRANSCRIPTOME ANALYSIS OF LAMINAR TISSUE DURING THE EARLY STAGES OF EXPERIMENTALLY INDUCED EQUINE LAMINITIS A Dissertation by JIXIN WANG Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY December 2010 Major Subject: Biomedical Sciences GENOME-WIDE TRANSCRIPTOME ANALYSIS OF LAMINAR TISSUE DURING THE EARLY STAGES OF EXPERIMENTALLY INDUCED EQUINE LAMINITIS A Dissertation by JIXIN WANG Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Approved by: Chair of Committee, Bhanu P. Chowdhary Committee Members, Terje Raudsepp Paul B. Samollow Loren C. Skow Penny K. Riggs Head of Department, Evelyn Tiffany-Castiglioni December 2010 Major Subject: Biomedical Sciences iii ABSTRACT Genome-wide Transcriptome Analysis of Laminar Tissue During the Early Stages of Experimentally Induced Equine Laminitis. (December 2010) Jixin Wang, B.S., Tarim University of Agricultural Reclamation; M.S., South China Agricultural University; M.S., Texas A&M University Chair of Advisory Committee: Dr. Bhanu P. Chowdhary Equine laminitis is a debilitating disease that causes extreme sufferring in afflicted horses and often results in a lifetime of chronic pain. The exact sequence of pathophysiological events culminating in laminitis has not yet been characterized, and this is reflected in the lack of any consistently effective therapeutic strategy. For these reasons, we used a newly developed 21,000 element equine-specific whole-genome oligoarray to perform transcriptomic analysis on laminar tissue from horses with experimentally induced models of laminitis: carbohydrate overload (CHO), hyperinsulinaemia (HI), and oligofructose (OF). -

WO 2019/068007 Al Figure 2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/068007 Al 04 April 2019 (04.04.2019) W 1P O PCT (51) International Patent Classification: (72) Inventors; and C12N 15/10 (2006.01) C07K 16/28 (2006.01) (71) Applicants: GROSS, Gideon [EVIL]; IE-1-5 Address C12N 5/10 (2006.0 1) C12Q 1/6809 (20 18.0 1) M.P. Korazim, 1292200 Moshav Almagor (IL). GIBSON, C07K 14/705 (2006.01) A61P 35/00 (2006.01) Will [US/US]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., C07K 14/725 (2006.01) P.O. Box 4044, 7403635 Ness Ziona (TL). DAHARY, Dvir [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (21) International Application Number: Box 4044, 7403635 Ness Ziona (IL). BEIMAN, Merav PCT/US2018/053583 [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (22) International Filing Date: Box 4044, 7403635 Ness Ziona (E.). 28 September 2018 (28.09.2018) (74) Agent: MACDOUGALL, Christina, A. et al; Morgan, (25) Filing Language: English Lewis & Bockius LLP, One Market, Spear Tower, SanFran- cisco, CA 94105 (US). (26) Publication Language: English (81) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of national protection available): AE, AG, AL, AM, 62/564,454 28 September 2017 (28.09.2017) US AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, 62/649,429 28 March 2018 (28.03.2018) US CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (71) Applicant: IMMP ACT-BIO LTD.