Anatomical Profiling of G Protein-Coupled Receptor Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Evidence for Gliadin Antibodies As Causative Agents in Schizophrenia
1 Evidence for gliadin antibodies as causative agents in schizophrenia. C.J.Carter PolygenicPathways, 20 Upper Maze Hill, Saint-Leonard’s on Sea, East Sussex, TN37 0LG [email protected] Tel: 0044 (0)1424 422201 I have no fax Abstract Antibodies to gliadin, a component of gluten, have frequently been reported in schizophrenia patients, and in some cases remission has been noted following the instigation of a gluten free diet. Gliadin is a highly immunogenic protein, and B cell epitopes along its entire immunogenic length are homologous to the products of numerous proteins relevant to schizophrenia (p = 0.012 to 3e-25). These include members of the DISC1 interactome, of glutamate, dopamine and neuregulin signalling networks, and of pathways involved in plasticity, dendritic growth or myelination. Antibodies to gliadin are likely to cross react with these key proteins, as has already been observed with synapsin 1 and calreticulin. Gliadin may thus be a causative agent in schizophrenia, under certain genetic and immunological conditions, producing its effects via antibody mediated knockdown of multiple proteins relevant to the disease process. Because of such homology, an autoimmune response may be sustained by the human antigens that resemble gliadin itself, a scenario supported by many reports of immune activation both in the brain and in lymphocytes in schizophrenia. Gluten free diets and removal of such antibodies may be of therapeutic benefit in certain cases of schizophrenia. 2 Introduction A number of studies from China, Norway, and the USA have reported the presence of gliadin antibodies in schizophrenia 1-5. Gliadin is a component of gluten, intolerance to which is implicated in coeliac disease 6. -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

Mechanisms of Acetylcholine Receptor Loss in Myasthenia Gravis
J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp.43.7.601 on 1 July 1980. Downloaded from Journal of Neurology, Neurosurgery, and Psychiatry, 1980, 43, 601-610 Mechanisms of acetylcholine receptor loss in myasthenia gravis DANIEL B DRACHMAN, ROBERT N ADAMS, ELIS F STANLEY, AND ALAN PESTRONK From the Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA SUMMARY The fundamental abnormality affecting the neuromuscular junctions of myasthenic patients is a reduction of available AChRs, due to an autoimmune attack directed against the receptors. Antibodies to AChR are present in most patients, and there is evidence that they have a predominant pathogenic role in the disease, aided by complement. The mechanism of antibody action involves acceleration of the rate of degradation of AChRs, attributable to cross-linking of the receptors. In addition, antibodies may block AChRs, and may participate in producing destructive changes, perhaps in conjunction with complement. The possibility that cell-mediated mechanisms may play a role in the autoimmune responses of some myasthenic patients remains to be explored. Although the target of the autoimmune attack in myasthenic patients is probably always the acetyl- Protected by copyright. choline receptors, it is not yet clear which of these immune mechanisms are most important. It is likely that the relative role of each mechanism varies from patient to patient. One of the goals of future research will be to identify the relative importance of each -

Edinburgh Research Explorer
Edinburgh Research Explorer International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list Citation for published version: Davenport, AP, Alexander, SPH, Sharman, JL, Pawson, AJ, Benson, HE, Monaghan, AE, Liew, WC, Mpamhanga, CP, Bonner, TI, Neubig, RR, Pin, JP, Spedding, M & Harmar, AJ 2013, 'International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein-coupled receptor list: recommendations for new pairings with cognate ligands', Pharmacological reviews, vol. 65, no. 3, pp. 967-86. https://doi.org/10.1124/pr.112.007179 Digital Object Identifier (DOI): 10.1124/pr.112.007179 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Pharmacological reviews Publisher Rights Statement: U.S. Government work not protected by U.S. copyright General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 02. Oct. 2021 1521-0081/65/3/967–986$25.00 http://dx.doi.org/10.1124/pr.112.007179 PHARMACOLOGICAL REVIEWS Pharmacol Rev 65:967–986, July 2013 U.S. -

High Throughput Circrna Sequencing Analysis Reveals Novel Insights Into
Xiong et al. Cell Death and Disease (2019) 10:658 https://doi.org/10.1038/s41419-019-1890-9 Cell Death & Disease ARTICLE Open Access High throughput circRNA sequencing analysis reveals novel insights into the mechanism of nitidine chloride against hepatocellular carcinoma Dan-dan Xiong1, Zhen-bo Feng1, Ze-feng Lai2,YueQin2, Li-min Liu3,Hao-xuanFu2, Rong-quan He4,Hua-yuWu5, Yi-wu Dang1, Gang Chen 1 and Dian-zhong Luo1 Abstract Nitidine chloride (NC) has been demonstrated to have an anticancer effect in hepatocellular carcinoma (HCC). However, the mechanism of action of NC against HCC remains largely unclear. In this study, three pairs of NC-treated and NC-untreated HCC xenograft tumour tissues were collected for circRNA sequencing analysis. In total, 297 circRNAs were differently expressed between the two groups, with 188 upregulated and 109 downregulated, among which hsa_circ_0088364 and hsa_circ_0090049 were validated by real-time quantitative polymerase chain reaction. The in vitro experiments showed that the two circRNAs inhibited the malignant biological behaviour of HCC, suggesting that they may play important roles in the development of HCC. To elucidate whether the two circRNAs function as “miRNA sponges” in HCC, we identified circRNA-miRNA and miRNA-mRNA interactions by using the CircInteractome and miRwalk, respectively. Subsequently, 857 miRNA-associated differently expressed genes in HCC were selected for weighted gene co-expression network analysis. Module Eigengene turquoise with 423 genes was found to be significantly related to the survival time, pathology grade and TNM stage of HCC patients. Gene functional enrichment 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; analysis showed that the 423 genes mainly functioned in DNA replication- and cell cycle-related biological processes and signalling cascades. -

Supplementary Data
Supplemental Data A novel mouse model of X-linked nephrogenic diabetes insipidus: Phenotypic analysis and therapeutic implications Jian Hua Li, Chung-Lin Chou, Bo Li, Oksana Gavrilova, Christoph Eisner, Jürgen Schnermann, Stasia A. Anderson, Chu-Xia Deng, Mark A. Knepper, and Jürgen Wess Supplemental Methods Metabolic cage studies. Animals were maintained in mouse metabolic cages (Hatteras Instruments, Cary, NC) under controlled temperature and light conditions (12 hr light and dark cycles). Mice received a fixed daily ration of 6.5 g of gelled diet per 20 g of body weight per day. The gelled diet was composed of 4 g of Basal Diet 5755 (Test Diet, Richmond, IN), 2.5 ml of deionized water, and 65 mg agar. Preweighted drinking water was provided ad libitum during the course of the study. Mice were acclimated in the metabolic cages for 1-2 days. Urine was collected under mineral oil in preweighted collection vials for successive 24 hr periods. Analysis of GPCR expression in mouse IMCD cells via TaqMan real-time qRT-PCR. Total RNA prepared from mouse IMCD tubule suspensions was reverse transcribed as described under Experimental Procedures. Tissues from ten 10-week old C57BL/6 WT mice were collected and pooled for each individual experiment. cDNA derived from 640 ng of RNA was mixed with an equal volume of TaqMan gene expression 2 x master mix (Applied Biosystems, Foster City, CA). 100 μl-aliquots of this mixture (corresponding to 80 ng of RNA) were added to each of the 8 fill ports of a 384-well plate of a mouse GPCR array panel (Applied Biosystems). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Protease Effects on the Structure of Acetylcholine Receptor Membranes from Torpedo Californica
PROTEASE EFFECTS ON THE STRUCTURE OF ACETYLCHOLINE RECEPTOR MEMBRANES FROM TORPEDO CALIFORNICA MICHAEL W. KLYMKOWSKY, JOHN E . HEUSER, and ROBERT M. STROUD From the Department of Biochemistry & Biophysics, University of California at San Francisco, San Francisco, California 94143 . Dr . Klymkowsky's present address is MRC Neuroimmunology Project, Department of Zoology, University College London, London WC IE, 6BT, England ABSTRACT Protease digestion of acetylcholine receptor-rich membranes derived from Torpedo californica electroplaques by homogenization and isopycnic centrifugation results in degradation of all receptor subunits without any significant effect on the appearance in electron micrographs, the toxin binding ability, or the sedimentation value of the receptor molecule . Such treatment does produce dramatic changes in the morphology of the normally 0.5- to 2-lm-diameter spherical vesicles when observed by either negative-stain or freeze-fracture electron microscopy . Removal of peripheral, apparently nonreceptor polypeptides by alkali stripping (Neubig et al ., 1979, Proc. Natl. Acad. Sci. U. S. A. 76:690-694) results in increased sensitivity of the acetylcholine receptor membranes to the protease trypsin as indicated by SDS gel electrophoretic patterns and by the extent of morphologic change observed in vesicle structure . Trypsin digestion of alkali-stripped receptor membranes results in a limit degradation pattern of all four receptor subunits, whereupon all the vesicles undergo the morphological transformation to minivesicles -

Interplay Between Gating and Block of Ligand-Gated Ion Channels
brain sciences Review Interplay between Gating and Block of Ligand-Gated Ion Channels Matthew B. Phillips 1,2, Aparna Nigam 1 and Jon W. Johnson 1,2,* 1 Department of Neuroscience, University of Pittsburgh, Pittsburgh, PA 15260, USA; [email protected] (M.B.P.); [email protected] (A.N.) 2 Center for Neuroscience, University of Pittsburgh, Pittsburgh, PA 15260, USA * Correspondence: [email protected]; Tel.: +1-(412)-624-4295 Received: 27 October 2020; Accepted: 26 November 2020; Published: 1 December 2020 Abstract: Drugs that inhibit ion channel function by binding in the channel and preventing current flow, known as channel blockers, can be used as powerful tools for analysis of channel properties. Channel blockers are used to probe both the sophisticated structure and basic biophysical properties of ion channels. Gating, the mechanism that controls the opening and closing of ion channels, can be profoundly influenced by channel blocking drugs. Channel block and gating are reciprocally connected; gating controls access of channel blockers to their binding sites, and channel-blocking drugs can have profound and diverse effects on the rates of gating transitions and on the stability of channel open and closed states. This review synthesizes knowledge of the inherent intertwining of block and gating of excitatory ligand-gated ion channels, with a focus on the utility of channel blockers as analytic probes of ionotropic glutamate receptor channel function. Keywords: ligand-gated ion channel; channel block; channel gating; nicotinic acetylcholine receptor; ionotropic glutamate receptor; AMPA receptor; kainate receptor; NMDA receptor 1. Introduction Neuronal information processing depends on the distribution and properties of the ion channels found in neuronal membranes. -

Characterization of the Lung Seven Transmembrane Recptor Protein
Florida State University Libraries Electronic Theses, Treatises and Dissertations The Graduate School 2012 Characterization of the Lung Seven Transmembrane (LUSTR) Protein Family Jennifer Ivey Middlebrooks Follow this and additional works at the FSU Digital Library. For more information, please contact [email protected] THE FLORIDA STATE UNIVERSITY COLLEGE OF ARTS AND SCIENCES CHARACTERIZATION OF THE LUNG SEVEN TRANSMEMBRANE (LUSTR) PROTEIN FAMILY By JENNIFER IVEY MIDDLEBROOKS A Dissertation submitted to the Department of Biological Sciences in partial fulfillment of the requirements for the degree of Doctor of Philosophy Degree Awarded: Summer Semester, 2012 Jennifer Ivey Middlebrooks defended this dissertation on June 1, 2012. The members of the supervisory committee were: Laura R. Keller Professor Directing Dissertation Cathy Levenson University Representative Tom Keller Committee Member Karen McGinnis Committee Member Wumin Deng Committee Member The Graduate School has verified and approved the above-named committee members, and certifies that the dissertation has been approved in accordance with university requirements. ii This work is dedicated to my loving, supportive family. iii ACKNOWLEDGEMENTS We thank Dr. Kenneth Roux and Leanna Willison (FSU Biological Science Dept.) for technical advice and reagents for completion of the immunoblot analysis, Dr. Tim MeGraw (FSU College of Medicine) for the use of his reagents and equipment for the immunofluorescent imaging, Dr. Gavin J.P. Naylor (College of Charleston) and Dr. Steven M. Thompson (South Georgia College) for their instruction and advice on the bioinformatic and phylogenetic analysis, and we acknowledge the Florida State University shared High-Performance Computing facility for facilitating the bioinformatic analyses. Additionally, we thank Dr. Joan Hare for her direction with human cell culture, Cody DeBrabaunt for his assistance in cell staining and microscopy, and Dr. -
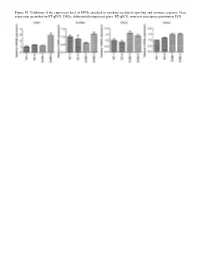
Figure S1. Validation of the Expression Level of Degs Enriched in Cytokine‑Mediated Signaling and Immune Response
Figure S1. Validation of the expression level of DEGs enriched in cytokine‑mediated signaling and immune response. Gene expression quantified by RT‑qPCR. DEGs, differentially expressed genes; RT‑qPCR, reverse‑transcription quantitative PCR. Figure S2. Validation of STAU1‑regulated AS events. IGV‑Sashimi plot revealed (A‑C) three A5SS AS events in three different genes. Reads distribution of each AS event was plotted in the left panel with the transcripts of each gene shown below. The sche‑ matic diagrams depict the structures of two AS events, AS1 (purple line) and AS2 (green line). The exon sequences are denoted by black boxes, the intron sequences by a horizontal line (right panel). RNA‑seq quantification and RT‑qPCR validation of ASEs are presented in the panels on the right. STAU1, double‑stranded RNA‑binding protein Staufen homolog 1; AS, alternative splicing; A5SS, alternative 5'splice site; RNA‑seq, RNA sequencing; RT‑qPCR, reverse‑transcription quantitative PCR. Error bars represent mean ± SEM. *P<0.05. Table SI. Primers used in gene validation experiments. IFIT2‑F CAGCCTACGGCAACTAAA IFIT2‑R GAGCCTTCTCAAAGCACA IFIT3‑F ACACCAAACAATGGCTAC IFIT3‑R TGGACAAACCCTCTAAAC OASL‑F AATGGTGACCGTGATGGG OASL‑R ACCTGAGGATGGAGCAGAG IFI27‑F TTCACTGCGGCGGGAATC IFI27‑R TGGCTGCTATGGAGGACGAG S1PR4‑F TGCTGAAGACGGTGCTGATG S1PR4‑R TGCGGAAGGAGTAGATGATGG CCL5‑F ACGACTGCTGGGTTGGAG CCL5‑R ACCCTGCTGCTTTGCCTA CCL2‑F CTAACCCAGAAACATCCAAT CCL2‑R GCTATGAGCAGCAGGCAC CD44‑F TGGAGGACAGAAAGCCAAGT CD44‑R TTCGCAATGAAACAATCAGTAG PLEKHG2‑M/As‑F CCAAAAGTAAGCCTGTCC PLEKHG2‑M‑R -

Synaptamide Activates the Adhesion GPCR GPR110 (ADGRF1) Through GAIN Domain Binding
ARTICLE https://doi.org/10.1038/s42003-020-0831-6 OPEN Synaptamide activates the adhesion GPCR GPR110 (ADGRF1) through GAIN domain binding Bill X. Huang1, Xin Hu2, Heung-Sun Kwon1, Cheng Fu1, Ji-Won Lee1, Noel Southall2, Juan Marugan2 & ✉ Hee-Yong Kim1 1234567890():,; Adhesion G protein-coupled receptors (aGPCR) are characterized by a large extracellular region containing a conserved GPCR-autoproteolysis-inducing (GAIN) domain. Despite their relevance to several disease conditions, we do not understand the molecular mechanism by which aGPCRs are physiologically activated. GPR110 (ADGRF1) was recently deorphanized as the functional receptor of N-docosahexaenoylethanolamine (synaptamide), a potent synap- togenic metabolite of docosahexaenoic acid. Thus far, synaptamide is the first and only small- molecule endogenous ligand of an aGPCR. Here, we demonstrate the molecular basis of synaptamide-induced activation of GPR110 in living cells. Using in-cell chemical cross-linking/ mass spectrometry, computational modeling and mutagenesis-assisted functional assays, we discover that synaptamide specifically binds to the interface of GPR110 GAIN subdomains through interactions with residues Q511, N512 and Y513, causing an intracellular conforma- tional change near TM6 that triggers downstream signaling. This ligand-induced GAIN-tar- geted activation mechanism provides a framework for understanding the physiological function of aGPCRs and therapeutic targeting in the GAIN domain. 1 Laboratory of Molecular Signaling, National Institute on Alcohol Abuse