Common Variants in Mendelian Kidney Disorder Genes and Their Association with Renal Function And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
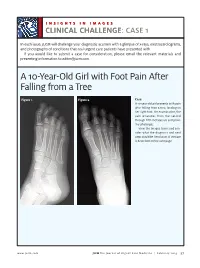
A 10-Year-Old Girl with Foot Pain After Falling from a Tree
INSIGHTS IN IMAGES CLINICAL CHALLENGECHALLENGE: CASE 1 In each issue, JUCM will challenge your diagnostic acumen with a glimpse of x-rays, electrocardiograms, and photographs of conditions that real urgent care patients have presented with. If you would like to submit a case for consideration, please email the relevant materials and presenting information to [email protected]. A 10-Year-Old Girl with Foot Pain After Falling from a Tree Figure 1. Figure 2. Case A 10-year-old girl presents with pain after falling from a tree, landing on her right foot. On examination, the pain emanates from the second through fifth metatarsals and proxi- mal phalanges. View the images taken and con- sider what the diagnosis and next steps would be. Resolution of the case is described on the next page. www.jucm.com JUCM The Journal of Urgent Care Medicine | February 2019 37 INSIGHTS IN IMAGES: CLINICAL CHALLENGE THE RESOLUTION Figure 1. Mediastinal air Figure 1. Differential Diagnosis Pearls for Urgent Care Management and Ⅲ Fracture of the distal fourth metatarsal Considerations for Transfer Ⅲ Plantar plate disruption Ⅲ Emergent transfer should be considered with associated neu- Ⅲ Sesamoiditis rologic deficit, compartment syndrome, open fracture, or vas- Ⅲ Turf toe cular compromise Ⅲ Referral to an orthopedist is warranted in the case of an in- Diagnosis tra-articular fracture, or with Lisfranc ligament injury or ten- Angulation of the distal fourth metatarsal metaphyseal cortex derness over the Lisfranc ligament and hairline lucency consistent with fracture. Acknowledgment: Images courtesy of Teleradiology Associates. Learnings/What to Look for Ⅲ Proximal metatarsal fractures are most often caused by crush- ing or direct blows Ⅲ In athletes, an axial load placed on a plantar-flexed foot should raise suspicion of a Lisfranc injury 38 JUCM The Journal of Urgent Care Medicine | February 2019 www.jucm.com INSIGHTS IN IMAGES CLINICAL CHALLENGE: CASE 2 A 55-Year-Old Man with 3 Hours of Epigastric Pain 55 years PR 249 QRSD 90 QT 471 QTc 425 AXES P 64 QRS -35 T 30 Figure 1. -

PERK Antibody / EIF2AK3 (RQ4206)
PERK Antibody / EIF2AK3 (RQ4206) Catalog No. Formulation Size RQ4206 0.5mg/ml if reconstituted with 0.2ml sterile DI water 100 ug Bulk quote request Availability 1-3 business days Species Reactivity Human, Mouse, Rat Format Antigen affinity purified Clonality Polyclonal (rabbit origin) Isotype Rabbit IgG Purity Antigen affinity purified Buffer Lyophilized from 1X PBS with 2% Trehalose and 0.025% sodium azide UniProt Q9NZJ5 Applications Western Blot : 0.5-1ug/ml Flow cytometry : 1-3ug/10^6 cells Direct ELISA : 0.1-0.5ug/ml Limitations This PERK antibody is available for research use only. Western blot testing of human 1) HeLa, 2) COLO320, 3) A549, 4) SK-OV-3, 5) A431, 6) rat brain and 7) mouse brain lysate with PERK antibody at 0.5ug/ml. Predicted molecular weight ~125 kDa, observed here at ~140 kDa. Flow cytometry testing of human HepG2 cells with PERK antibody at 1ug/10^6 cells (blocked with goat sera); Red=cells alone, Green=isotype control, Blue= PERK antibody. Description Eukaryotic translation initiation factor 2-alpha kinase 3, also known as protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK), is an enzyme that in humans is encoded by the EIF2AK3 gene. The protein encoded by this gene phosphorylates the alpha subunit of eukaryotic translation-initiation factor 2, leading to its inactivation, and thus to a rapid reduction of translational initiation and repression of global protein synthesis. This protein is thought to modulate mitochondrial function. It is a type I membrane protein located in the endoplasmic reticulum (ER), where it is induced by ER stress caused by malfolded proteins. -

Leading Article the Molecular and Genetic Base of Congenital Transport
Gut 2000;46:585–587 585 Gut: first published as 10.1136/gut.46.5.585 on 1 May 2000. Downloaded from Leading article The molecular and genetic base of congenital transport defects In the past 10 years, several monogenetic abnormalities Given the size of SGLT1 mRNA (2.3 kb), the gene is large, have been identified in families with congenital intestinal with 15 exons, and the introns range between 3 and 2.2 kb. transport defects. Wright and colleagues12 described the A single base change was identified in the entire coding first, which concerns congenital glucose and galactose region of one child, a finding that was confirmed in the malabsorption. Subsequently, altered genes were identified other aZicted sister. This was a homozygous guanine to in partial or total loss of nutrient absorption, including adenine base change at position 92. The patient’s parents cystinuria, lysinuric protein intolerance, Menkes’ disease were heterozygotes for this mutation. In addition, it was (copper malabsorption), bile salt malabsorption, certain found that the 92 mutation was associated with inhibition forms of lipid malabsorption, and congenital chloride diar- of sugar transport by the protein. Since the first familial rhoea. Altered genes may also result in decreased secretion study, genomic DNA has been screened in 31 symptomatic (for chloride in cystic fibrosis) or increased absorption (for GGM patients in 27 kindred from diVerent parts of the sodium in Liddle’s syndrome or copper in Wilson’s world. In all 33 cases the mutation produced truncated or disease)—for general review see Scriver and colleagues,3 mutant proteins. -

Next Generation Sequencing Panels for Disorders of Sex Development
Next Generation Sequencing Panels for Disorders of Sex Development Disorders of Sex Development – Overview Disorders of sex development (DSDs) occur when sex development does not follow the course of typical male or female patterning. Types of DSDs include congenital development of ambiguous genitalia, disjunction between the internal and external sex anatomy, incomplete development of the sex anatomy, and abnormalities of the development of gonads (such as ovotestes or streak ovaries) (1). Sex chromosome anomalies including Turner syndrome and Klinefelter syndrome as well as sex chromosome mosaicism are also considered to be DSDs. DSDs can be caused by a wide range of genetic abnormalities (2). Determining the etiology of a patient’s DSD can assist in deciding gender assignment, provide recurrence risk information for future pregnancies, and can identify potential health problems such as adrenal crisis or gonadoblastoma (1, 3). Sex chromosome aneuploidy and copy number variation are common genetic causes of DSDs. For this reason, chromosome analysis and/or microarray analysis typically should be the first genetic analysis in the case of a patient with ambiguous genitalia or other suspected disorder of sex development. Identifying whether a patient has a 46,XY or 46,XX karyotype can also be helpful in determining appropriate additional genetic testing. Abnormal/Ambiguous Genitalia Panel Our Abnormal/Ambiguous Genitalia Panel includes mutation analysis of 72 genes associated with both syndromic and non-syndromic DSDs. This comprehensive panel evaluates a broad range of genetic causes of ambiguous or abnormal genitalia, including conditions in which abnormal genitalia are the primary physical finding as well as syndromic conditions that involve abnormal genitalia in addition to other congenital anomalies. -

Genetic Mechanisms of Disease in Children: a New Look
Genetic Mechanisms of Disease in Children: A New Look Laurie Demmer, MD Tufts Medical Center and the Floating Hospital for Children Boston, MA Traditionally genetic disorders have been linked to the ‘one gene-one protein-one disease’ hypothesis. However recent advances in the field of molecular biology and biotechnology have afforded us the opportunity to greatly expand our knowledge of genetics, and we now know that the mechanisms of inherited disorders are often significantly more complex, and consequently, much more intriguing, than originally thought. Classical mendelian disorders with relatively simple genetic mechanisms do exist, but turn out to be far more rare than originally thought. All patients with sickle cell disease for example, carry the same A-to-T point mutation in the sixth codon of the beta globin gene. This results in a glutamate to valine substitution which changes the shape and the function of the globin molecule in a predictable way. Similarly, all patients with achondroplasia have a single base pair substitution at nucleotide #1138 of the FGFR3 gene. On the other hand, another common inherited disorder, cystic fibrosis, is known to result from changes in a specific transmembrane receptor (CFTR), but over 1000 different disease-causing mutations have been reported in this single gene. Since most commercial labs only test for between 23-100 different mutations, interpreting CFTR mutation testing is significantly complicated by the known risk of false negative results. Many examples of complex, or non-Mendelian, inheritance are now known to exist and include disorders of trinucleotide repeats, errors in imprinting, and gene dosage effects. -
Second-Tier DNA Confirmation of Newborn Screening Results
Second-tier DNA Confirmation of Newborn Screening by Targeted Next Generation Sequencing Edwin Naylor, Ph.D. MPH Andy Bhattacharjee , Ph.D. Erik Puffenberger, Ph.D.; Kevin Strauss, MD; Holmes Morton, MD Newborn Screening & Clinical Genomics 1961 1990’s 2010-2012 2 Robert Guthrie Development of develops simple automated MS/MS Newborn Screening screening across (NBS) several disorders Current de facto standard 2 Why Newborn Genomics? • Mendelian Diseases disproportionately affect Newborns - ~3500 genetic diseases with molecular basis - >10% of NICU admissions are genetic Clinical manifestation of Genetic diseases - Current NBS tests limited to 29+ diseases CHROMOSOMAL - 2nd tier DNA testing to validate biochemical results MULTI-FACTORIAL SINGLE GENE (MENDELIAN) • Advantage of NGS based DNA testing individuals # of Affected - Find causal variants (rare/novel) in gene(s) - A ‘universal’ NGS approach avoids repeated, serial BIRTH PUBERTY ADULT single gene testing Gelehrter TD, Collins FS, Ginsburg D. Principles of - Current Sanger sequencing is expensive ($3-10K) and Medical Genetics. 2nd ed. Baltimore, MD: Williams & slow (3 months to 1 year) Wilkins; 1998:1-42 NICU- Neonatal Intensive Care Unit NBS-Newborn Screening 3 NGS-Next Generation Sequencing Why Targeted (Exome) Sequencing for now? NGS Sequencing * Genomic DNA from Causal Mutations in Affected Individuals Exons/Target Regions Fold Test Menu Cost ($) Throughput Efficiency Whole Genome (Res.) 7,666* 1 1 Exome (Res) 1,200 15 95 Neonate Panel (Clinical) <1000 150 >1140 •Majority of known disease-causing mutations in exons •Exome = protein-encoding parts of genes •Targeted NGS is Cost & Throughput Efficient *Saunders et al., (2012) Rapid Whole Genome Sequencing for Genetic Disease Diagnosis in NICUs 4 Workflow for 2nd Tier Newborn Screening Sample 2h DNA Capture 92h Raw Data 10h Analysis 1h+ Isolation & Sequencing Management & Interpretation 8 samples, 105 Hrs, <$10,000 = Real Neonatal Genomics! 5 Workflow for 2nd Tier Newborn Screening Sample •High M.Wt. -

Supplement 1 Overview of Dystonia Genes
Supplement 1 Overview of genes that may cause dystonia in children and adolescents Gene (OMIM) Disease name/phenotype Mode of inheritance 1: (Formerly called) Primary dystonias (DYTs): TOR1A (605204) DYT1: Early-onset generalized AD primary torsion dystonia (PTD) TUBB4A (602662) DYT4: Whispering dystonia AD GCH1 (600225) DYT5: GTP-cyclohydrolase 1 AD deficiency THAP1 (609520) DYT6: Adolescent onset torsion AD dystonia, mixed type PNKD/MR1 (609023) DYT8: Paroxysmal non- AD kinesigenic dyskinesia SLC2A1 (138140) DYT9/18: Paroxysmal choreoathetosis with episodic AD ataxia and spasticity/GLUT1 deficiency syndrome-1 PRRT2 (614386) DYT10: Paroxysmal kinesigenic AD dyskinesia SGCE (604149) DYT11: Myoclonus-dystonia AD ATP1A3 (182350) DYT12: Rapid-onset dystonia AD parkinsonism PRKRA (603424) DYT16: Young-onset dystonia AR parkinsonism ANO3 (610110) DYT24: Primary focal dystonia AD GNAL (139312) DYT25: Primary torsion dystonia AD 2: Inborn errors of metabolism: GCDH (608801) Glutaric aciduria type 1 AR PCCA (232000) Propionic aciduria AR PCCB (232050) Propionic aciduria AR MUT (609058) Methylmalonic aciduria AR MMAA (607481) Cobalamin A deficiency AR MMAB (607568) Cobalamin B deficiency AR MMACHC (609831) Cobalamin C deficiency AR C2orf25 (611935) Cobalamin D deficiency AR MTRR (602568) Cobalamin E deficiency AR LMBRD1 (612625) Cobalamin F deficiency AR MTR (156570) Cobalamin G deficiency AR CBS (613381) Homocysteinuria AR PCBD (126090) Hyperphelaninemia variant D AR TH (191290) Tyrosine hydroxylase deficiency AR SPR (182125) Sepiaterine reductase -

Inherited Metabolic Disease
Inherited metabolic disease Dr Neil W Hopper SRH Areas for discussion • Introduction to IEMs • Presentation • Initial treatment and investigation of IEMs • Hypoglycaemia • Hyperammonaemia • Other presentations • Management of intercurrent illness • Chronic management Inherited Metabolic Diseases • Result from a block to an essential pathway in the body's metabolism. • Huge number of conditions • All rare – very rare (except for one – 1:500) • Presentation can be non-specific so index of suspicion important • Mostly AR inheritance – ask about consanguinity Incidence (W. Midlands) • Amino acid disorders (excluding phenylketonuria) — 18.7 per 100,000 • Phenylketonuria — 8.1 per 100,000 • Organic acidemias — 12.6 per 100,000 • Urea cycle diseases — 4.5 per 100,000 • Glycogen storage diseases — 6.8 per 100,000 • Lysosomal storage diseases — 19.3 per 100,000 • Peroxisomal disorders — 7.4 per 100,000 • Mitochondrial diseases — 20.3 per 100,000 Pathophysiological classification • Disorders that result in toxic accumulation – Disorders of protein metabolism (eg, amino acidopathies, organic acidopathies, urea cycle defects) – Disorders of carbohydrate intolerance – Lysosomal storage disorders • Disorders of energy production, utilization – Fatty acid oxidation defects – Disorders of carbohydrate utilization, production (ie, glycogen storage disorders, disorders of gluconeogenesis and glycogenolysis) – Mitochondrial disorders – Peroxisomal disorders IMD presentations • ? IMD presentations • Screening – MCAD, PKU • Progressive unexplained neonatal -

The Genetic Basis for Skeletal Diseases
insight review articles The genetic basis for skeletal diseases Elazar Zelzer & Bjorn R. Olsen Harvard Medical School, Department of Cell Biology, 240 Longwood Avenue, Boston, Massachusetts 02115, USA (e-mail: [email protected]) We walk, run, work and play, paying little attention to our bones, their joints and their muscle connections, because the system works. Evolution has refined robust genetic mechanisms for skeletal development and growth that are able to direct the formation of a complex, yet wonderfully adaptable organ system. How is it done? Recent studies of rare genetic diseases have identified many of the critical transcription factors and signalling pathways specifying the normal development of bones, confirming the wisdom of William Harvey when he said: “nature is nowhere accustomed more openly to display her secret mysteries than in cases where she shows traces of her workings apart from the beaten path”. enetic studies of diseases that affect skeletal differentiation to cartilage cells (chondrocytes) or bone cells development and growth are providing (osteoblasts) within the condensations. Subsequent growth invaluable insights into the roles not only of during the organogenesis phase generates cartilage models individual genes, but also of entire (anlagen) of future bones (as in limb bones) or membranous developmental pathways. Different mutations bones (as in the cranial vault) (Fig. 1). The cartilage anlagen Gin the same gene may result in a range of abnormalities, are replaced by bone and marrow in a process called endo- and disease ‘families’ are frequently caused by mutations in chondral ossification. Finally, a process of growth and components of the same pathway. -

Renal Agenesis, Renal Tubular Dysgenesis, and Polycystic Renal Diseases
Developmental & Structural Anomalies of the Genitourinary Tract DR. Alao MA Bowen University Teach Hosp Ogbomoso Picture test Introduction • Congenital Anomalies of the Kidney & Urinary Tract (CAKUT) Objectives • To review the embryogenesis of UGS and dysmorphogenesis of CAKUT • To describe the common CAKUT in children • To emphasize the role of imaging in the diagnosis of CAKUT Introduction •CAKUT refers to gross structural anomalies of the kidneys and or urinary tract present at birth. •Malformation of the renal parenchyma resulting in failure of normal nephron development as seen in renal dysplasia, renal agenesis, renal tubular dysgenesis, and polycystic renal diseases. Introduction •Abnormalities of embryonic migration of the kidneys as seen in renal ectopy (eg, pelvic kidney) and fusion anomalies, such as horseshoe kidney. •Abnormalities of the developing urinary collecting system as seen in duplicate collecting systems, posterior urethral valves, and ureteropelvic junction obstruction. Introduction •Prevalence is about 3-6 per 1000 births •CAKUT is one of the commonest anomalies found in human. •It constitute approximately 20 to 30 percent of all anomalies identified in the prenatal period •The presence of CAKUT in a child raises the chances of finding congenital anomalies of other organ-systems Why the interest in CAKUT? •Worldwide, CAKUT plays a causative role in 30 to 50 percent of cases of end-stage renal disease (ESRD), •The presence of CAKUT, especially ones affecting the bladder and lower tract adversely affects outcome of kidney graft after transplantation Why the interest in CAKUT? •They significantly predispose the children to UTI and urinary calculi •They may be the underlying basis for urinary incontinence Genes & Environment Interact to cause CAKUT? • Tens of different genes with role in nephrogenesis have been identified. -

De Novo EIF2AK1 and EIF2AK2 Variants Are Associated with Developmental Delay, Leukoencephalopathy, and Neurologic Decompensation
bioRxiv preprint doi: https://doi.org/10.1101/757039; this version posted September 16, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. De novo EIF2AK1 and EIF2AK2 variants are associated with developmental delay, leukoencephalopathy, and neurologic decompensation Dongxue Mao1,2, Chloe M. Reuter3,4, Maura R.Z. Ruzhnikov5,6, Anita E. Beck7, Emily G. Farrow8,9,10, Lisa T. Emrick1,11,12,13, Jill A. Rosenfeld12, Katherine M. Mackenzie5, Laurie Robak2,12,13, Matthew T. Wheeler3,14, Lindsay C. Burrage12,13, Mahim Jain15, Pengfei Liu12, Daniel Calame11,13, Sebastien Küry17,18, Martin Sillesen19, Klaus Schmitz-Abe20, Davide Tonduti21, Luigina Spaccini22, Maria Iascone23, Casie A. Genetti20, Madeline Graf16, Alyssa Tran12, Mercedes Alejandro12, Undiagnosed Diseases Network, Brendan H. Lee12,13, Isabelle Thiffault8,9,24, Pankaj B. Agrawal#,20, Jonathan A. Bernstein#,3,25, Hugo J. Bellen#,2,12,26,27,28, Hsiao- Tuan Chao#,1,2,11,12,13,28,27,29 #Correspondence should be addressed: [email protected] (P.A.), [email protected] (J.A.B.), [email protected] (H.J.B.), [email protected] (H.T.C.) 1Department of Pediatrics, Baylor College of Medicine (BCM), Houston, TX 2Jan and Dan Duncan Neurological Research Institute, Texas Children’s Hospital, Houston, TX 3Stanford Center for Undiagnosed Diseases, Stanford University, Stanford, CA 4Stanford Center for Inherited Cardiovascular Disease, Division of Cardiovascular Medicine, -

Congenital Disorders of Glycosylation from a Neurological Perspective
brain sciences Review Congenital Disorders of Glycosylation from a Neurological Perspective Justyna Paprocka 1,* , Aleksandra Jezela-Stanek 2 , Anna Tylki-Szyma´nska 3 and Stephanie Grunewald 4 1 Department of Pediatric Neurology, Faculty of Medical Science in Katowice, Medical University of Silesia, 40-752 Katowice, Poland 2 Department of Genetics and Clinical Immunology, National Institute of Tuberculosis and Lung Diseases, 01-138 Warsaw, Poland; [email protected] 3 Department of Pediatrics, Nutrition and Metabolic Diseases, The Children’s Memorial Health Institute, W 04-730 Warsaw, Poland; [email protected] 4 NIHR Biomedical Research Center (BRC), Metabolic Unit, Great Ormond Street Hospital and Institute of Child Health, University College London, London SE1 9RT, UK; [email protected] * Correspondence: [email protected]; Tel.: +48-606-415-888 Abstract: Most plasma proteins, cell membrane proteins and other proteins are glycoproteins with sugar chains attached to the polypeptide-glycans. Glycosylation is the main element of the post- translational transformation of most human proteins. Since glycosylation processes are necessary for many different biological processes, patients present a diverse spectrum of phenotypes and severity of symptoms. The most frequently observed neurological symptoms in congenital disorders of glycosylation (CDG) are: epilepsy, intellectual disability, myopathies, neuropathies and stroke-like episodes. Epilepsy is seen in many CDG subtypes and particularly present in the case of mutations