Detecting Ulcerative Colitis from Colon Samples Using Efficient Feature
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Unusual Presentation of a Young Child with Cirrhosis: a Case Report
Central Annals of Pediatrics & Child Health Case Report *Corresponding author Sina Aziz, Abbasi Shaheed Hospital and Karachi Medical and Dental College, Block M, North Unusual Presentation of a Nazimabad, Karachi 74700, Pakistan, Tel: 03008213278; Email: Submitted: 07 March 2015 Young Child with Cirrhosis: A Accepted: 21 April 2015 Published: 23 April 2015 Case Report Copyright © 2015 Aziz et al. Sundus Khan1, Shahameen Aqeel1, Muhammad Arif2 and Sina Aziz3* OPEN ACCESS 1 Medical Student, Karachi Medical and Dental College, Pakistan Keywords 2 Department of Pathology, Karachi Medical and Dental College, Pakistan • Cirrhosis 3 Department of Pediatrics, Abbasi Shaheed Hospital and Karachi Medical and Dental • Hepatitis College, Pakistan • Biliary atresia • Glycogen storage disease Abstract • Alpha-1 antitrypsin deficiency • Tyrosinemia Cirrhosis (Greek word) by definition is a hard, nodular regenerating disease of • Cystic fibrosis liver in which the hepatocytes are constantly injured (by insulting agent) with fibrosis due to increase in connective tissues that ultimately lead to destruction in structure and function of liver. Cirrhosis in paediatric population occur due to acute/chronic liver damage which may be due to viral hepatitis (HBV, HBV and HDV co infection, HCV, CMV or NANB hepatitis), autoimmune disorders, toxins (drug induced), certain inborn errors of metabolism (Wilson`s disease, alpha-1 antitrypsin deficiency, tyrosinemiaetc.), glycogen storage disease or cryptogenic.We report a case of a 2 year 6 month old baby boy who presented with bleeding from nose, abdominal distension and hepatomegaly; investigation revealed a cirrhotic liver disease pattern. INTRODUCTION intermittent nasal bleeding since one year, abdominal distension since one year and freshnasal bleed inthe last one-week. -

Inflammatory Bowel Disease Irritable Bowel Syndrome
Inflammatory Bowel Disease and Irritable Bowel Syndrome Similarities and Differences 2 www.ccfa.org IBD Help Center: 888.MY.GUT.PAIN 888.694.8872 Important Differences Between IBD and IBS Many diseases and conditions can affect the gastrointestinal (GI) tract, which is part of the digestive system and includes the esophagus, stomach, small intestine and large intestine. These diseases and conditions include inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS). IBD Help Center: 888.MY.GUT.PAIN 888.694.8872 www.ccfa.org 3 Inflammatory bowel diseases are a group of inflammatory conditions in which the body’s own immune system attacks parts of the digestive system. Inflammatory Bowel Disease Inflammatory bowel diseases are a group of inflamma- Causes tory conditions in which the body’s own immune system attacks parts of the digestive system. The two most com- The exact cause of IBD remains unknown. Researchers mon inflammatory bowel diseases are Crohn’s disease believe that a combination of four factors lead to IBD: a (CD) and ulcerative colitis (UC). IBD affects as many as 1.4 genetic component, an environmental trigger, an imbal- million Americans, most of whom are diagnosed before ance of intestinal bacteria and an inappropriate reaction age 35. There is no cure for IBD but there are treatments to from the immune system. Immune cells normally protect reduce and control the symptoms of the disease. the body from infection, but in people with IBD, the immune system mistakes harmless substances in the CD and UC cause chronic inflammation of the GI tract. CD intestine for foreign substances and launches an attack, can affect any part of the GI tract, but frequently affects the resulting in inflammation. -

Nutritional Considerations in Inflammatory Bowel Disease
NUTRITION ISSUES IN GASTROENTEROLOGY, SERIES #5 Series Editor: Carol Rees Parrish, M.S., R.D., CNSD Nutritional Considerations in Inflammatory Bowel Disease by Kelly Anne Eiden, M.S., R.D., CNSD Nutrient alterations are commonplace in patients with inflammatory bowel disease. The etiology for these alterations is multifactorial. Nutrition assessment is the first step in successful nutrition management of any patient with gastrointestinal disease. Nutritional goals include assisting with nutrition risk, identifying macronutrient and micronutrient needs and implementing a nutrition plan to meet those needs. This article addresses many of the nutrition issues currently facing clinicians including: oral, enteral and parenteral nutrition, common vitamin/mineral deficiencies, medium chain triglycerides and nutrition as primary and supportive therapy. INTRODUCTION and supportive treatment in both Crohn’s and UC. The nflammatory bowel disease (IBD), encompassing following article will provide guidelines to help the both Crohn’s disease and ulcerative colitis (UC), is clinician determine nutritional risk, review specialized Ia chronic inflammatory intestinal disorder of nutrient needs and discuss nutrition as a treatment unknown etiology. A multitude of factors, including modality in the patient with IBD. drug-nutrient interactions, disease location, symp- toms, and dietary restrictions can lead to protein NUTRITION ASSESSMENT IN INFLAMMATORY energy malnutrition and specific nutritional deficien- BOWEL DISEASE cies. It is estimated that up to 85% of hospitalized IBD patients have protein energy malnutrition, based on Factors Affecting Nutritional Status abnormal anthropometric and biochemical parameters in the Patient with IBD (1,2). As Crohn’s disease can occur anywhere from There are many factors that alter nutrient intake in the mouth to anus (80% of cases in the terminal ileum), it patient with IBD. -

Chronic Viral Hepatitis in a Cohort of Inflammatory Bowel Disease
pathogens Article Chronic Viral Hepatitis in a Cohort of Inflammatory Bowel Disease Patients from Southern Italy: A Case-Control Study Giuseppe Losurdo 1,2 , Andrea Iannone 1, Antonella Contaldo 1, Michele Barone 1 , Enzo Ierardi 1 , Alfredo Di Leo 1,* and Mariabeatrice Principi 1 1 Section of Gastroenterology, Department of Emergency and Organ Transplantation, University “Aldo Moro” of Bari, 70124 Bari, Italy; [email protected] (G.L.); [email protected] (A.I.); [email protected] (A.C.); [email protected] (M.B.); [email protected] (E.I.); [email protected] (M.P.) 2 Ph.D. Course in Organs and Tissues Transplantation and Cellular Therapies, Department of Emergency and Organ Transplantation, University “Aldo Moro” of Bari, 70124 Bari, Italy * Correspondence: [email protected]; Tel.: +39-080-559-2925 Received: 14 September 2020; Accepted: 21 October 2020; Published: 23 October 2020 Abstract: We performed an epidemiologic study to assess the prevalence of chronic viral hepatitis in inflammatory bowel disease (IBD) and to detect their possible relationships. Methods: It was a single centre cohort cross-sectional study, during October 2016 and October 2017. Consecutive IBD adult patients and a control group of non-IBD subjects were recruited. All patients underwent laboratory investigations to detect chronic hepatitis B (HBV) and C (HCV) infection. Parameters of liver function, elastography and IBD features were collected. Univariate analysis was performed by Student’s t or chi-square test. Multivariate analysis was performed by binomial logistic regression and odds ratios (ORs) were calculated. We enrolled 807 IBD patients and 189 controls. Thirty-five (4.3%) had chronic viral hepatitis: 28 HCV (3.4%, versus 5.3% in controls, p = 0.24) and 7 HBV (0.9% versus 0.5% in controls, p = 0.64). -

Liver Disease in Ulcerative Colitis: an Epidemiological and Follow up Study
84 Gut 1994; 35:84-89 Liver disease in ulcerative colitis: an epidemiological and follow up study in the county of Stockholm Gut: first published as 10.1136/gut.35.1.84 on 1 January 1994. Downloaded from U Broome, H Glaumann, G Hellers, B Nilsson, J Sorstad, R Hultcrantz Abstract sclerosing cholangitis (PSC) are said to be more In an epidemiological study ofthe incidence of common in patients with UC.47 The incidence of ulcerative colitis (UC) in the county of Stock- these complications varies in different studies holm between 1955 and 1979, 1274 patients depending on selection criteria and on the with UC were discovered. Almost all these definition of hepatobiliary disease.89 The true patients had regularly been investigated with incidence of hepatobiliary dysfunction in UC, liver function tests; 142 (11%) of them showed however, can only be obtained by tests of liver signs of hepatobiliary disease. A follow up function tests such as transaminases, alkaline study on all 142 patients with abnormal liver phosphatases, and bilirubin made routinely in function and UC was made between 1989 and unselected groups of patients with UC. Because 1991 to evaluate the cause of the liver abnor- the cause of hepatic involvement in UC is mality and to find out if the liver disease had enigmatic it is important to find out if the rate affected the survival rates. At follow up, eight of this complication is increasing, because of patients were reclassified as having Crohn's factors unrelated to UC, or if it mirrors the disease, 60 had developed normal liver func- incidence ofUC in itself. -

C4BQ0: a Genetic Marker of Familial HCV-Related Liver Cirrhosis
Digestive and Liver Disease 36 (2004) 471–477 Liver, Pancreas and Biliary Tract C4BQ0: a genetic marker of familial HCV-related liver cirrhosis L. Pasta b,∗, G. Pietrosi c, C. Marrone a, G. D’Amico b, M. D’Amico a, A. Licata d, G. Misiano e, S. Madonia b, F. Mercadante a, L. Pagliaro a,b a Department of Medicine and Pneumology, “V Cervello” Hospital, Via Trabucco 180, 90146 Palermo, Italy b CRRMCF (Centro di Riferimento Regionale delle Malattie Croniche di Fegato), Division of Medicine, “V Cervello” Hospital, Via Trabucco 180, 90146 Palermo, Italy c Department of Medicine and Gastroenetrology, IsMeTT-UPMC, Palermo, Italy d Gastroenterology Unit, Department of Internal Medicine, Palermo, Italy e Department of Immunology, University of Palermo, Palermo, Italy Received 7 August 2003; accepted 2 February 2004 Available online 5 May 2004 Abstract Background and methods. Host may have a role in the evolution of chronic HCV liver disease. We performed two cross-sectional prospective studies to evaluate the prevalence of cirrhosis in first degree relatives of patients with cirrhosis and the role of two major histocompatibility complex class III alleles BF and C4 versus HCV as risk factors for familial clustering. Findings. Ninety-three (18.6%) of 500 patients with cirrhosis had at least one cirrhotic first degree relative as compared to 13 (2.6%) of 500 controls, (OR 7.38; CI 4.21–12.9). C4BQ0 was significantly more frequent in the 93 cirrhotic patients than in 93 cirrhotic controls without familiarity (Hardy–Weinberg equilibrium: 2 5.76, P = 0.016) and in 20 families with versus 20 without aggregation of HCV related cirrhosis (29.2% versus 11.3%, P = 0.001); the association C4BQ0-HCV was found almost only in cirrhotic patients with a family history of liver cirrhosis. -

Progress Report Genetics and Gastroenterology
Gut: first published as 10.1136/gut.12.7.592 on 1 July 1971. Downloaded from Gut, 1971, 12, 592-598 Progress report Genetics and gastroenterology The decade since the last review of this subject in Gut has seen considerable advances in many aspects of genetics, including the influence of heredity on disease. Compared with some specialties gastroenterology has few disorders with simple Mendelian inheritance, but in most of its conditions there is some hereditary influence. Some of the best human data illustrating a quantitative type of disease liability are those on hypertrophic pyloric stenosis and Hirschsprung's disease, and the discovery of two of the genes of the genotype giving susceptibility to duodenal ulcer is still unique. There is growing realization that both heredity and environmental factors are concerned in the aetiology of most disorders. Blood Groups and Alimentary Physiology It is still not clear what part the ABO blood group genes and the genes at the secretor, H, and Lewis loci play in alimentary physiology1. It would seem likely that they are concerned via their control of the arrangement of the sugars on the molecules of the mucus of the upper part of the tract. Yet one would not expect such slight variations between the different group specific http://gut.bmj.com/ molecules to affect their physicochemical properties. Furthermore, the dis- covery of the close relationship between the serum content ofjejunal alkaline phosphatase and the ABO blood group and secretor status2,3,4 suggests that the blood group genes are pleiotropic. In addition to their well known, and possibly unimportant, effect on the serological specificity of glycoproteins, these genes may be concerned in more fundamental processes such as the on October 2, 2021 by guest. -
Ulcerative Colitis Your Guide
CROHN’S & COLITIS UK SUPPORTING YOU TO MANAGE YOUR CONDITION ULCERATIVE COLITIS YOUR GUIDE 3 INTRODUCTION ABOUT THIS BOOKLET Being diagnosed with Ulcerative Colitis can be a shock, and you probably have many questions. Now that you’ve put a name to your symptoms, you can start to manage them. The more you know about your Colitis, the more you’ll be able to take an active part in decisions about your care. This booklet will give you and your family and friends a better understanding of Colitis, how it is treated, and how it will affect your life. All our publications are research-based and produced in consultation with patients, medical advisers and other health or associated professionals. They are prepared as general information and are not intended to replace specific advice from your own doctor or any other professional. Crohn’s & Colitis UK does not endorse or recommend any products mentioned. If you would like more information about the sources of evidence on which this booklet is based, or details of any conflicts of interest, or if you have any feedback on our publications, please email publications@ crohnsandcolitis.org.uk. About Crohn’s & Colitis UK We are a national charity established in 1979. Our aim is to improve life for anyone affected by Crohn’s and Colitis. We have 40,000 members and 50 Local Networks throughout the UK. Membership costs start from £15 per year with concessionary rates for anyone experiencing financial hardship or on a low income. © Crohn’s & Colitis UK 2017 & 2019 This publication is available free of charge, but we Ulcerative Colitis Edition 9d would not be able to do this without our supporters and Last review - June 2017 members. -

Fact Sheet: News from the IBD Help Center: Intestinal Complications
Fact Sheet News from the IBD Help Center INTESTINAL COMPLICATIONS The complications of Crohn’s disease and ulcerative colitis are generally classified as either local or systemic. The term “local” refers to complications involving the intestinal tract itself, while the term “systemic” (or extraintestinal) refers to complications that involve other organs or that affect the patient as a whole. Intestinal complications tend to occur when the intestinal inflammation: • is severe • extends beyond the inner lining (mucosa) of the intestines • is widespread • is chronic (of long duration) Some intestinal complications occur in both ulcerative colitis and Crohn’s disease, although they may occur more commonly in one than in the other. Not everyone will experience these complications. However, early recognition and prompt treatment are key. If you notice a change in your symptoms, be sure to contact your doctor immediately. Ulcerative Colitis-Local Complications Perforation (rupture) of the bowel. Intestinal perforation occurs when chronic inflammation and ulceration of the intestine weakens the wall to such an extent that a hole develops in the intestinal wall. This perforation is potentially life- threatening because the contents of the intestine, which contain a large number of bacteria, can spill into the abdomen and cause a serious infection called peritonitis. In colitis, this complication is generally linked with toxic megacolon (see below). In Crohn’s disease, it may occur as a result of an abscess or fistula. Fulminant colitis. This complication, which affects less than 10% of people with colitis, involves damage to the entire thickness of the intestinal wall. When severe inflammation causes the colon to become extremely dilated and swollen, a condition called ileus may develop. -

Enteric Viruses and Inflammatory Bowel Disease
viruses Review Enteric Viruses and Inflammatory Bowel Disease Georges Tarris 1,2, Alexis de Rougemont 2 , Maëva Charkaoui 3, Christophe Michiels 3, Laurent Martin 1 and Gaël Belliot 2,* 1 Department of Pathology, University Hospital of Dijon, F 21000 Dijon, France; [email protected] (G.T.); [email protected] (L.M.) 2 National Reference Centre for Gastroenteritis Viruses, Laboratory of Virology, University Hospital of Dijon, F 21000 Dijon, France; [email protected] 3 Department of Hepatogastroenterology, University Hospital of Dijon, F 21000 Dijon, France; [email protected] (M.C.); [email protected] (C.M.) * Correspondence: [email protected]; Tel.: +33-380-293-171; Fax: +33-380-293-280 Abstract: Inflammatory bowel diseases (IBD), including ulcerative colitis (UC) and Crohn’s disease (CD), is a multifactorial disease in which dietary, genetic, immunological, and microbial factors are at play. The role of enteric viruses in IBD remains only partially explored. To date, epidemiological studies have not fully described the role of enteric viruses in inflammatory flare-ups, especially that of human noroviruses and rotaviruses, which are the main causative agents of viral gastroenteritis. Genome-wide association studies have demonstrated the association between IBD, polymorphisms of the FUT2 and FUT3 genes (which drive the synthesis of histo-blood group antigens), and ligands for norovirus and rotavirus in the intestine. The role of autophagy in defensin-deficient Paneth cells and the perturbations of cytokine secretion in T-helper 1 and T-helper 17 inflammatory pathways following enteric virus infections have been demonstrated as well. -
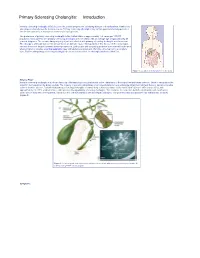
Primary Sclerosing Cholangitis: Introduction
Primary Sclerosing Cholangitis: Introduction Primary sclerosing cholangitis (PSC) is a chronic , usually progressive, stricturing disease of the biliary tree. Remissions and relapses characterize the disease course. Primary sclerosing cholangitis may remain quiescent for long periods of time in some patients; in most cases, however, it is progressive. The prevalence of primary sclerosing cholangitis in the United States is approximately 1–6 cases per 100,000 population. Most patients with primary sclerosing cholangitis are men (75%) with an average age of approximately 40 years at diagnosis. The overwhelming majority of patients affected with primary sclerosing cholangitis are Caucasian. The etiology is unknown but current opinion favors an immune cause. Management of this disease in the early stages involves the use of drugs to prevent disease progression. Endoscopic and surgical approaches are reserved for the time when symptoms develop. Liver transplantation may ultimately be required and offers the only chance for a complete cure. Patients with primary sclerosing cholangitis are at an increased risk for cholangiocarcinoma (10–15%). Figure 1. Location of the biliary tree in the body. What is PSC? Primary sclerosing cholangitis is a chronic fibrosing inflammatory process that results in the obliteration of the biliary tree and biliary cirrhosis. There is variability in the extent of involvement of the biliary system. The majority of patients with primary sclerosing cholangitis have underlying inflammatory bowel disease, namely ulcerative colitis or Crohn’s disease. Patients with primary sclerosing cholangitis are more likely to have ulcerative colitis than Crohn’s disease (85% versus 15%), with approximately 2.5–7.5% of all ulcerative colitis patients having primary sclerosing cholangitis. -

The Risk of Colorectal Cancer in Patients with Ulcerative Colitis
Dig Dis Sci (2015) 60:492–501 DOI 10.1007/s10620-014-3373-2 ORIGINAL ARTICLE The Risk of Colorectal Cancer in Patients with Ulcerative Colitis Tobias M. Nowacki • Markus Bru¨ckner • Maria Eveslage • Phil Tepasse • Friederike Pott • Nils H. Thoennissen • Karin Hengst • Matthias Ross • Dominik Bettenworth Received: 11 April 2014 / Accepted: 24 September 2014 / Published online: 4 October 2014 Ó Springer Science+Business Media New York 2014 Abstract associated with the risk of CAC (P \ 0.0014); disease Background and Aim Ulcerative colitis increases the risk duration between 9 and 15 years: OR of 2.5 (95 % CI of developing dysplasia and colitis-associated cancer 0.2–41.1), more than 15 years: OR of 21.4 (95 % CI (CAC). The purpose of this study was to determine the risk 2.6–173.6). The risk of developing dysplasia (low-grade factors as well as protective measures for disease burden, intraepithelial neoplasia, LGIEN and high-grade intraepi- need for colectomy and the development of CAC in thelial neoplasia, HGIEN) or the need to undergo colec- ulcerative colitis (UC) patients. tomy was also significantly related to disease duration Methods A cohort of n = 434 UC patients was evaluated. (P = 0.006, P = 0.002, respectively). Established anti- Data analysis was performed by univariate and multivariate inflammatory medication (e.g., 5-ASA, anti-TNF-a) sig- logistic regression. Odds ratios (OR) and 95 % confidence nificantly reduced the risk of both dysplasia and CAC intervals (CI) were calculated, and significance was (P = 0.02). assessed by the likelihood ratio test. Conclusions Despite the use of modern therapies for UC, Results Mean patient age at UC diagnosis was CAC rates remain high.