An Introduction to the Psych Package: Part I: Data Entry and Data Description
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Psychometric Analysis of the Flow Short Scale Translated to Finnish
Psychometric Analysis of the Flow Short Scale translated to Finnish Michael Laakasuoa, Jussi Palomäkia, Sami Abuhamdehb, Otto Lappia,c and Benjamin Ultan Cowleya,d aCognitive Science, Department of Digital Humanities, Faculty of Arts, University of Helsinki; Siltavuorenpenger 1 A, 00012 Helsinki, Finland bDepartment of Psychology, Marmara University, Istanbul, Turkey cTraffic Research Unit, University of Helsinki, Helsinki, Finland dFaculty of Educational Sciences, University of Helsinki, Helsinki, Finland ARTICLEINFO ABSTRACT Keywords: We report a psychometric scale validation study on a Finnish translation of the Flow Short Scale Flow (FSS). Data from 201 Finnish speaking participants were collected via the Prolific Academic Finnish Flow Short Scale platform. To analyse the scale’s factorial structure, we performed Mokken scale analysis, Par- Scale validation allel Analysis, Very Simple Structures analysis and a standard Confirmatory Factor Analysis. Psychometrics Then, we evaluated the correlations between FSS and two other flow scales, the Flow State Scale and Flow Core Scale. Finally, we evaluated the FSS’s ability to distinguish flow-inducing experiences from boring (non-flow-inducing) experiences. Collectively the results showed that an 8-item, two-factor version of the scale was a justified instrument with good psychometric properties. 1. Introduction Flow is a well-known construct describing the experience of harmony between action and awareness, typically in a demanding but intrinsically-motivating task conducted with high skill [4]. As an experiential construct, Flow can only be assessed by some form of self-report. Various instruments have been developed to thus operationalise Flow [7, 12, 15], but these have not been made available in a range of languages in thoroughly validated form. -

Synthpop: Bespoke Creation of Synthetic Data in R
synthpop: Bespoke Creation of Synthetic Data in R Beata Nowok Gillian M Raab Chris Dibben University of Edinburgh University of Edinburgh University of Edinburgh Abstract In many contexts, confidentiality constraints severely restrict access to unique and valuable microdata. Synthetic data which mimic the original observed data and preserve the relationships between variables but do not contain any disclosive records are one possible solution to this problem. The synthpop package for R, introduced in this paper, provides routines to generate synthetic versions of original data sets. We describe the methodology and its consequences for the data characteristics. We illustrate the package features using a survey data example. Keywords: synthetic data, disclosure control, CART, R, UK Longitudinal Studies. This introduction to the R package synthpop is a slightly amended version of Nowok B, Raab GM, Dibben C (2016). synthpop: Bespoke Creation of Synthetic Data in R. Journal of Statistical Software, 74(11), 1-26. doi:10.18637/jss.v074.i11. URL https://www.jstatsoft. org/article/view/v074i11. 1. Introduction and background 1.1. Synthetic data for disclosure control National statistical agencies and other institutions gather large amounts of information about individuals and organisations. Such data can be used to understand population processes so as to inform policy and planning. The cost of such data can be considerable, both for the collectors and the subjects who provide their data. Because of confidentiality constraints and guarantees issued to data subjects the full access to such data is often restricted to the staff of the collection agencies. Traditionally, data collectors have used anonymisation along with simple perturbation methods such as aggregation, recoding, record-swapping, suppression of sensitive values or adding random noise to prevent the identification of data subjects. -

Bootstrapping Regression Models
Bootstrapping Regression Models Appendix to An R and S-PLUS Companion to Applied Regression John Fox January 2002 1 Basic Ideas Bootstrapping is a general approach to statistical inference based on building a sampling distribution for a statistic by resampling from the data at hand. The term ‘bootstrapping,’ due to Efron (1979), is an allusion to the expression ‘pulling oneself up by one’s bootstraps’ – in this case, using the sample data as a population from which repeated samples are drawn. At first blush, the approach seems circular, but has been shown to be sound. Two S libraries for bootstrapping are associated with extensive treatments of the subject: Efron and Tibshirani’s (1993) bootstrap library, and Davison and Hinkley’s (1997) boot library. Of the two, boot, programmed by A. J. Canty, is somewhat more capable, and will be used for the examples in this appendix. There are several forms of the bootstrap, and, additionally, several other resampling methods that are related to it, such as jackknifing, cross-validation, randomization tests,andpermutation tests. I will stress the nonparametric bootstrap. Suppose that we draw a sample S = {X1,X2, ..., Xn} from a population P = {x1,x2, ..., xN };imagine further, at least for the time being, that N is very much larger than n,andthatS is either a simple random sample or an independent random sample from P;1 I will briefly consider other sampling schemes at the end of the appendix. It will also help initially to think of the elements of the population (and, hence, of the sample) as scalar values, but they could just as easily be vectors (i.e., multivariate). -

A Survey on Data Collection for Machine Learning a Big Data - AI Integration Perspective
1 A Survey on Data Collection for Machine Learning A Big Data - AI Integration Perspective Yuji Roh, Geon Heo, Steven Euijong Whang, Senior Member, IEEE Abstract—Data collection is a major bottleneck in machine learning and an active research topic in multiple communities. There are largely two reasons data collection has recently become a critical issue. First, as machine learning is becoming more widely-used, we are seeing new applications that do not necessarily have enough labeled data. Second, unlike traditional machine learning, deep learning techniques automatically generate features, which saves feature engineering costs, but in return may require larger amounts of labeled data. Interestingly, recent research in data collection comes not only from the machine learning, natural language, and computer vision communities, but also from the data management community due to the importance of handling large amounts of data. In this survey, we perform a comprehensive study of data collection from a data management point of view. Data collection largely consists of data acquisition, data labeling, and improvement of existing data or models. We provide a research landscape of these operations, provide guidelines on which technique to use when, and identify interesting research challenges. The integration of machine learning and data management for data collection is part of a larger trend of Big data and Artificial Intelligence (AI) integration and opens many opportunities for new research. Index Terms—data collection, data acquisition, data labeling, machine learning F 1 INTRODUCTION E are living in exciting times where machine learning expertise. This problem applies to any novel application that W is having a profound influence on a wide range of benefits from machine learning. -

Three Statistical Testing Procedures in Logistic Regression: Their Performance in Differential Item Functioning (DIF) Investigation
Research Report Three Statistical Testing Procedures in Logistic Regression: Their Performance in Differential Item Functioning (DIF) Investigation Insu Paek December 2009 ETS RR-09-35 Listening. Learning. Leading.® Three Statistical Testing Procedures in Logistic Regression: Their Performance in Differential Item Functioning (DIF) Investigation Insu Paek ETS, Princeton, New Jersey December 2009 As part of its nonprofit mission, ETS conducts and disseminates the results of research to advance quality and equity in education and assessment for the benefit of ETS’s constituents and the field. To obtain a PDF or a print copy of a report, please visit: http://www.ets.org/research/contact.html Copyright © 2009 by Educational Testing Service. All rights reserved. ETS, the ETS logo, GRE, and LISTENING. LEARNING. LEADING. are registered trademarks of Educational Testing Service (ETS). SAT is a registered trademark of the College Board. Abstract Three statistical testing procedures well-known in the maximum likelihood approach are the Wald, likelihood ratio (LR), and score tests. Although well-known, the application of these three testing procedures in the logistic regression method to investigate differential item function (DIF) has not been rigorously made yet. Employing a variety of simulation conditions, this research (a) assessed the three tests’ performance for DIF detection and (b) compared DIF detection in different DIF testing modes (targeted vs. general DIF testing). Simulation results showed small differences between the three tests and different testing modes. However, targeted DIF testing consistently performed better than general DIF testing; the three tests differed more in performance in general DIF testing and nonuniform DIF conditions than in targeted DIF testing and uniform DIF conditions; and the LR and score tests consistently performed better than the Wald test. -

William Revelle Department of Psychology Northwestern University 847-491-7700 [email protected]
William Revelle Department of Psychology Northwestern University 847-491-7700 [email protected] Education University of Michigan, Department of Psychology 1968-1973 Rackham Prize Fellow, 1970-1972 Ph.D. in Psychology received December, 1973 Pomona College: 1961-1965 B.A. in Psychology received June, 1965 Employment Northwestern University Chairman, Department of Psychology 1987–1993, 1998–2001 Professor of Psychology 1984 - present Associate Professor of Psychology 1979 - 1984 Assistant Professor of Psychology 1973 - 1979 University of Michigan Teaching Assistant, 1972-1973 Research Assistant, 1968-1971 United States Peace Corps Volunteer Sarawak, Malaysia 1965-1967 Professional Interests Psychometric theory and applications Personality theory and human motivation The effects of personality, motivational, and situational variables on cognitive processes Biological bases of affect, personality and behavior Google Scholar https://scholar.google.com/citations?user=293jbW0AAAAJ&hl=en 1 Revelle vita February, 2018 2 Honors and Awards President, International Society for the Study of Individual Differences 2005- 2009 President, Association for Research in Personality 2007-2009 President, Society of Multivariate Experimental Psychology, 1984-1985 Chair, Governing Board, Bulletin of Atomic Scientists, 2009 - 2012 Vice-Chair, Governing Board, Bulletin of Atomic Scientists, 2013 - present Board member, Bulletin of the Atomic Scientists, 2007 - present Fellow, American Association for the Advancement of Science, 1996-present Fellow, Association -
Testing for INAR Effects
Communications in Statistics - Simulation and Computation ISSN: 0361-0918 (Print) 1532-4141 (Online) Journal homepage: https://www.tandfonline.com/loi/lssp20 Testing for INAR effects Rolf Larsson To cite this article: Rolf Larsson (2020) Testing for INAR effects, Communications in Statistics - Simulation and Computation, 49:10, 2745-2764, DOI: 10.1080/03610918.2018.1530784 To link to this article: https://doi.org/10.1080/03610918.2018.1530784 © 2019 The Author(s). Published with license by Taylor & Francis Group, LLC Published online: 21 Jan 2019. Submit your article to this journal Article views: 294 View related articles View Crossmark data Full Terms & Conditions of access and use can be found at https://www.tandfonline.com/action/journalInformation?journalCode=lssp20 COMMUNICATIONS IN STATISTICS - SIMULATION AND COMPUTATIONVR 2020, VOL. 49, NO. 10, 2745–2764 https://doi.org/10.1080/03610918.2018.1530784 Testing for INAR effects Rolf Larsson Department of Mathematics, Uppsala University, Uppsala, Sweden ABSTRACT ARTICLE HISTORY In this article, we focus on the integer valued autoregressive model, Received 10 April 2018 INAR (1), with Poisson innovations. We test the null of serial independ- Accepted 25 September 2018 ence, where the INAR parameter is zero, versus the alternative of a KEYWORDS positive INAR parameter. To this end, we propose different explicit INAR model; Likelihood approximations of the likelihood ratio (LR) statistic. We derive the limit- ratio test ing distributions of our statistics under the null. In a simulation study, we compare size and power of our tests with the score test, proposed by Sun and McCabe [2013. Score statistics for testing serial depend- ence in count data. -

A First Step Into the Bootstrap World
INSTITUTE FOR DEFENSE ANALYSES A First Step into the Bootstrap World Matthew Avery May 2016 Approved for public release. IDA Document NS D-5816 Log: H 16-000624 INSTITUTE FOR DEFENSE ANALYSES 4850 Mark Center Drive Alexandria, Virginia 22311-1882 The Institute for Defense Analyses is a non-profit corporation that operates three federally funded research and development centers to provide objective analyses of national security issues, particularly those requiring scientific and technical expertise, and conduct related research on other national challenges. About This Publication This briefing motivates bootstrapping through an intuitive explanation of basic statistical inference, discusses the right way to resample for bootstrapping, and uses examples from operational testing where the bootstrap approach can be applied. Bootstrapping is a powerful and flexible tool when applied appropriately. By treating the observed sample as if it were the population, the sampling distribution of statistics of interest can be generated with precision equal to Monte Carlo error. This approach is nonparametric and requires only acceptance of the observed sample as an estimate for the population. Careful resampling is used to generate the bootstrap distribution. Applications include quantifying uncertainty for availability data, quantifying uncertainty for complex distributions, and using the bootstrap for inference, such as the two-sample t-test. Copyright Notice © 2016 Institute for Defense Analyses 4850 Mark Center Drive, Alexandria, Virginia 22311-1882 -
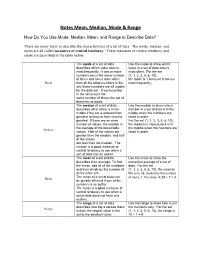
Notes Mean, Median, Mode & Range
Notes Mean, Median, Mode & Range How Do You Use Mode, Median, Mean, and Range to Describe Data? There are many ways to describe the characteristics of a set of data. The mode, median, and mean are all called measures of central tendency. These measures of central tendency and range are described in the table below. The mode of a set of data Use the mode to show which describes which value occurs value in a set of data occurs most frequently. If two or more most often. For the set numbers occur the same number {1, 1, 2, 3, 5, 6, 10}, of times and occur more often the mode is 1 because it occurs Mode than all the other numbers in the most frequently. set, those numbers are all modes for the data set. If each number in the set occurs the same number of times, the set of data has no mode. The median of a set of data Use the median to show which describes what value is in the number in a set of data is in the middle if the set is ordered from middle when the numbers are greatest to least or from least to listed in order. greatest. If there are an even For the set {1, 1, 2, 3, 5, 6, 10}, number of values, the median is the median is 3 because it is in the average of the two middle the middle when the numbers are Median values. Half of the values are listed in order. greater than the median, and half of the values are less than the median. -

Comparison of Wald, Score, and Likelihood Ratio Tests for Response Adaptive Designs
Journal of Statistical Theory and Applications Volume 10, Number 4, 2011, pp. 553-569 ISSN 1538-7887 Comparison of Wald, Score, and Likelihood Ratio Tests for Response Adaptive Designs Yanqing Yi1∗and Xikui Wang2 1 Division of Community Health and Humanities, Faculty of Medicine, Memorial University of Newfoundland, St. Johns, Newfoundland, Canada A1B 3V6 2 Department of Statistics, University of Manitoba, Winnipeg, Manitoba, Canada R3T 2N2 Abstract Data collected from response adaptive designs are dependent. Traditional statistical methods need to be justified for the use in response adaptive designs. This paper gener- alizes the Rao's score test to response adaptive designs and introduces a generalized score statistic. Simulation is conducted to compare the statistical powers of the Wald, the score, the generalized score and the likelihood ratio statistics. The overall statistical power of the Wald statistic is better than the score, the generalized score and the likelihood ratio statistics for small to medium sample sizes. The score statistic does not show good sample properties for adaptive designs and the generalized score statistic is better than the score statistic under the adaptive designs considered. When the sample size becomes large, the statistical power is similar for the Wald, the sore, the generalized score and the likelihood ratio test statistics. MSC: 62L05, 62F03 Keywords and Phrases: Response adaptive design, likelihood ratio test, maximum likelihood estimation, Rao's score test, statistical power, the Wald test ∗Corresponding author. Fax: 1-709-777-7382. E-mail addresses: [email protected] (Yanqing Yi), xikui [email protected] (Xikui Wang) Y. Yi and X. -

Robust Score and Portmanteau Tests of Volatility Spillover Mike Aguilar, Jonathan B
Journal of Econometrics 184 (2015) 37–61 Contents lists available at ScienceDirect Journal of Econometrics journal homepage: www.elsevier.com/locate/jeconom Robust score and portmanteau tests of volatility spillover Mike Aguilar, Jonathan B. Hill ∗ Department of Economics, University of North Carolina at Chapel Hill, United States article info a b s t r a c t Article history: This paper presents a variety of tests of volatility spillover that are robust to heavy tails generated by Received 27 March 2012 large errors or GARCH-type feedback. The tests are couched in a general conditional heteroskedasticity Received in revised form framework with idiosyncratic shocks that are only required to have a finite variance if they are 1 September 2014 independent. We negligibly trim test equations, or components of the equations, and construct heavy Accepted 1 September 2014 tail robust score and portmanteau statistics. Trimming is either simple based on an indicator function, or Available online 16 September 2014 smoothed. In particular, we develop the tail-trimmed sample correlation coefficient for robust inference, and prove that its Gaussian limit under the null hypothesis of no spillover has the same standardization JEL classification: C13 irrespective of tail thickness. Further, if spillover occurs within a specified horizon, our test statistics C20 obtain power of one asymptotically. We discuss the choice of trimming portion, including a smoothed C22 p-value over a window of extreme observations. A Monte Carlo study shows our tests provide significant improvements over extant GARCH-based tests of spillover, and we apply the tests to financial returns data. Keywords: Finally, based on ideas in Patton (2011) we construct a heavy tail robust forecast improvement statistic, Volatility spillover which allows us to demonstrate that our spillover test can be used as a model specification pre-test to Heavy tails Tail trimming improve volatility forecasting. -

Common Data Set 2020-2021 University of Pennsylvania
Common Data Set 2020-2021 University of Pennsylvania Table of Contents A General Information page 3 B Enrollment and Persistence 5 C First Time, First Year Admission 8 D Transfer Admission 14 E Academic Offerings and Policies 16 F Student Life 18 G Annual Expenses 20 H Financial Aid 22 I Instructional Faculty and Class Size 28 J Disciplinary Areas of Degrees Conferred 30 Common Data Set Definitions 32 Common Data Set 2020-21 1 25 Jun 2021 Common Data Set 2020-21 2 25 Jun 2021 A. General Information return to Table of Contents A1 Address Information A1 University of Pennsylvania A1 Mailing Address: 1 College Hall, Room 100 A1 City/State/Zip/Country: Philadelphia, PA 19104-6228 A1 Main Phone Number: 215-898-5000 A1 Home Page Address www.upenn.edu A1 Admissions Phone Number: 215-898-7507 A1 Admissions Office Mailing Address: 1 College Hall A1 City/State/Zip/Country: Philadelphia, PA 19104 A1 Admissions Fax Number: 215-898-9670 A1 Admissions E-mail Address: [email protected] A1 Online application www.admissions.upenn.edu A2 Source of institutional control (Check only one): A2 Public A2 Private (nonprofit) x A2 Proprietary A3 Classify your undergraduate institution: A3 Coeducational college A3 Men's college A3 Women's college A4 Academic year calendar: A4 Semester x A4 Quarter A4 Trimester A4 4-1-4 A4 Continuous A5 Degrees offered by your institution: A5 Certificate x A5 Diploma A5 Associate x A5 Transfer Associate A5 Terminal Associate x A5 Bachelor's x A5 Postbachelor's certificate x A5 Master's x A5 Post-master's certificate x A5 Doctoral degree - research/scholarship x A5 Doctoral degree - professional practice x A5 Doctoral degree - other x A5 Doctoral degree -- other Common Data Set 2020-21 3 25 Jun 2021 Common Data Set 2020-21 4 25 Jun 2021 B.