Master File IX
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Phosphorylation of Mcardle Phosphorylase Induces Activity (Human Skeletal Muscle/Protein Kinase) CESARE G
Proc. Nati. Acad. Sci. USA Vol. 78, No. 5, pp. 2688-2692, May 1981 Biochemistry Phosphorylation of McArdle phosphorylase induces activity (human skeletal muscle/protein kinase) CESARE G. CERRI AND JOSEPH H. WILLNER Department of Neurology and H. Houston Merritt Clinical Research Center for Muscular Dystrophy and Related Diseases, Columbia University College of Physicians and Surgeons, New York, New York 10032 Communicated by Harry Grundfest, January 7, 1981 ABSTRACT In McArdle disease, myophosphorylase defi- mediate between those of phosphorylases b and a. Karpatkin ciency, enzyme activity is absent but the presence of an altered et al. (19, 20) found that incubation of human platelets with enzyme protein can frequently be demonstrated. We have found MgATP+ resulted in an increase in total phosphorylase activity that phosphorylation of this protein in vitro can result in catalytic and concluded that the data were "consistent with the presence activity. We studied muscle of four patients; all lacked myophos- in human platelets of inactive dimer and monomer species of phorylase activity, but myophosphorylase protein was demon- phosphorylase, which require MgATP for activation." Because strated by immunodiffusion or gel electrophoresis. Incubation of activation of these isozymes was probably due to protein phos- muscle homogenate supernatants with cyclic AMP-dependent pro- phorylation and also because incomplete phosphorylation could tein kinase and ATP resulted in phosphorylase activity. The ac- tivated enzyme comigrated with normal human myophosphory- result in reduced activity, we evaluated the possibility that the lase in gel electrophoresis. Incubation with [y-32P]ATP resulted activity ofphosphorylase in McArdle muscle could be restored in incorporation of 32P into the band possessing phosphorylase by phosphorylation of the inactive phosphorylase protein pres- activity. -

Aliphatic 1-Amino Acid Decarboxylase from Ferns (Filicopsida) Thomas Hartmann
Aliphatic 1-Amino Acid Decarboxylase from Ferns (Filicopsida) Thomas Hartmann. Klaus Bax. and Renate Scholz Institut für Pharmazeutische Biologie der Technischen Universität Braunschweig, Mendels- sohnstr. 1, D-3300 Braunschweig. Bundesrepublik Deutschland Z. Naturforsch. 39c, 2 4 -3 0 (1984); received Septem ber 26, 1983 Ferns, Filicopsida, Polypodium vulgare. Aliphatic 1-Amino Acid Decarboxylase, Occurrence and Distribution A screening of 27 fern species (Filicopsida) out of 9 families revealed that 25 species were able to decarboxylate 1-leucine to 3-methylbutylamine (isoamylamine). The enzyme of Polvpodium vulgäre has partially been purified and characterisized. All attempts to solubilize it from acetone preparations failed; however, approx. 50% of total activity could be extracted from dry material in the presence of detergents at high concentration. The soluble enzyme was purified 132-fold. 1-Methionine was found the best substrate followed by norvaline, leucine, norleucine, isoleucine, homocysteine, valine. It has been confirmed that these substrates are decarboxylated by a single enzyme. The pH-optimum was at pH 5.0 (particulate preparation) and pH 4.5 (soluble enzyme). Decarboxylation is dependent on pyridoxal-5'-phosphate (PLP). A strictly substrate dependent coenzyme dissociation was observed which could largely be prevented by addition of 2 -oxo-acids, such as glyoxylate or pyruvate. Apodecarboxylase prepared by prolonged substrate incubation was found to be extremely labile at pH 4.5 but stable at pH 6.5. A comparison of the fern enzyme with bacterial valine decarboxylase (EC 4.1.1.14) and leucine decarboxylase of red algae revealed great similarities especially in substrate specificity. It is suggested to unify these activities as “aliphatic 1-amino acid decarboxylase”. -

Guaiacol As a Drug Candidate for Treating Adult Polyglucosan Body Disease
Guaiacol as a drug candidate for treating adult polyglucosan body disease Or Kakhlon, … , Wyatt W. Yue, H. Orhan Akman JCI Insight. 2018;3(17):e99694. https://doi.org/10.1172/jci.insight.99694. Research Article Metabolism Therapeutics Graphical abstract Find the latest version: https://jci.me/99694/pdf RESEARCH ARTICLE Guaiacol as a drug candidate for treating adult polyglucosan body disease Or Kakhlon,1 Igor Ferreira,2 Leonardo J. Solmesky,3 Netaly Khazanov,4 Alexander Lossos,1 Rafael Alvarez,5 Deniz Yetil,6 Sergey Pampou,7 Miguel Weil,3,8 Hanoch Senderowitz,4 Pablo Escriba,5 Wyatt W. Yue,2 and H. Orhan Akman9 1Department of Neurology, Hadassah-Hebrew University Medical Center, Jerusalem, Israel. 2Structural Genomics Consortium, Nuffield Department of Clinical Medicine, University of Oxford, Oxford, United Kingdom.3 Cell Screening Facility for Personalized Medicine, Department of Cell Research and Immunology, The George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel. 4Department of Chemistry, Bar Ilan University, Ramat Gan, Israel. 5Laboratory of Molecular Cell Biomedicine, Department of Biology, University of the Balearic Islands, Palma de Mallorca, Spain. 6Connecticut College, Newington, Connecticut USA. 7Columbia University Department of Systems Biology Irving Cancer Research Center, New York, New York, USA. 8Laboratory for Neurodegenerative Diseases and Personalized Medicine, Department of Cell Research and Immunology, The George S. Wise Faculty for Life Sciences, Sagol School of Neurosciences, Tel Aviv University, Ramat Aviv, Tel Aviv, Israel. 9Columbia University Medical Center Department of Neurology, Houston Merritt Neuromuscular diseases research center, New York, New York, USA. Adult polyglucosan body disease (APBD) is a late-onset disease caused by intracellular accumulation of polyglucosan bodies, formed due to glycogen-branching enzyme (GBE) deficiency. -

A Genome Based Discovery of S. Mansoni Secretome to Identify Therapeutic Targets
ics om & B te i ro o P in f f o o Mandage et al. J Proteomics Bioinform 2011, 4:3 r l m a Journal of a n t r i c u DOI: 10.4172/jpb.1000168 s o J ISSN: 0974-276X Proteomics & Bioinformatics Research Article Article OpenOpen Access Access A Genome Based Discovery of S. mansoni Secretome to Identify Therapeutic Targets Mandage RH1*, Jadhavrao PK2, Wadnerkar AS1, Varsale AR1 and Kurwade KK1 1Dept of Bioinformatics, Centre for Advanced Life Sciences, Deogiri College, Aurangabad 431005, M.S. India 2Dept of Microbiology, Abeda Inamdar College, Pune 411001, M.S. India Abstract The motivation behind the large scale genome analysis of S. mansoni was to explore the possibility of discovering the secretome that is frequently secreted at the interface of parasite and host is supposed to play a crucial role in parasitism by suppressing the immune response, to aid the proliferation of infectivity. Here, we present an efficient pipeline of bioinformatics methodology to identify candidate parasitism proteins within S. mansoni secretome of immune responses in the infected host. The 3,700 proteins deduced from the S. mansoni genome were analysed for the presence or absence of secretory signal peptides. We identified 32 proteins carrying an N-terminal secreted signal peptide but deficient in extra membrane-anchoring moieties. Notably we identified proteins involved in ATP synthesis, redox balance, protein folding, gluconeogenesis, development and signaling, scavenging and nucleotide metabolic pathways, immune response modulation. Most of these proteins define their potential for immunological diagnosis and vaccine design. A systematic attempt has been made here to develop a general method for predicting secretory proteins of a parasite with high efficiency and accuracy. -

An Evolving Hierarchical Family Classification for Glycosyltransferases
doi:10.1016/S0022-2836(03)00307-3 J. Mol. Biol. (2003) 328, 307–317 COMMUNICATION An Evolving Hierarchical Family Classification for Glycosyltransferases Pedro M. Coutinho1, Emeline Deleury1, Gideon J. Davies2 and Bernard Henrissat1* 1Architecture et Fonction des Glycosyltransferases are a ubiquitous group of enzymes that catalyse the Macromole´cules Biologiques transfer of a sugar moiety from an activated sugar donor onto saccharide UMR6098, CNRS and or non-saccharide acceptors. Although many glycosyltransferases catalyse Universite´s d’Aix-Marseille I chemically similar reactions, presumably through transition states with and II, 31 Chemin Joseph substantial oxocarbenium ion character, they display remarkable diversity Aiguier, 13402 Marseille in their donor, acceptor and product specificity and thereby generate a Cedex 20, France potentially infinite number of glycoconjugates, oligo- and polysacchar- ides. We have performed a comprehensive survey of glycosyltransferase- 2Structural Biology Laboratory related sequences (over 7200 to date) and present here a classification of Department of Chemistry, The these enzymes akin to that proposed previously for glycoside hydrolases, University of York, Heslington into a hierarchical system of families, clans, and folds. This evolving York YO10 5YW, UK classification rationalises structural and mechanistic investigation, harnesses information from a wide variety of related enzymes to inform cell biology and overcomes recurrent problems in the functional prediction of glycosyltransferase-related open-reading frames. q 2003 Elsevier Science Ltd. All rights reserved Keywords: glycosyltransferases; protein families; classification; modular *Corresponding author structure; genomic annotations The biosynthesis of complex carbohydrates and are involved in glycosyl transfer and thus conquer polysaccharides is of remarkable biological import- what Sharon has provocatively described as “the ance. -
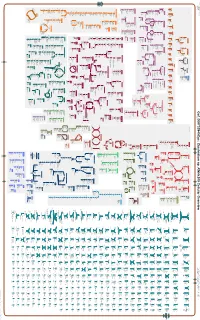
Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000702945Cyc: Clostridium sp. KNHs205 Cellular Overview Connections between pathways are omitted for legibility. -

Chem331 Glycogen Metabolism
Glycogen metabolism Glycogen review - 1,4 and 1,6 α-glycosidic links ~ every 10 sugars are branched - open helix with many non-reducing ends. Effective storage of glucose Glucose storage Liver glycogen 4.0% 72 g Muscle glycogen 0.7% 245 g Blood Glucose 0.1% 10 g Large amount of water associated with glycogen - 0.5% of total weight Glycogen stored in granules in cytosol w/proteins for synthesis, degradation and control There are very different means of control of glycogen metabolism between liver and muscle Glycogen biosynthetic and degradative cycle Two different pathways - which do not share enzymes like glycolysis and gluconeogenesis glucose -> glycogen glycogenesis - biosynthetic glycogen -> glucose 1-P glycogenolysis - breakdown Evidence for two paths - Patients lacking phosphorylase can still synthesize glycogen - hormonal regulation of both directions Glycogenolysis (glycogen breakdown)- Glycogen Phosphorylase glycogen (n) + Pi -> glucose 1-p + glycogen (n-1) • Enzyme binds and cleaves glycogen into monomers at the end of the polymer (reducing ends of glycogen) • Dimmer interacting at the N-terminus. • rate limiting - controlled step in glycogen breakdown • glycogen phosphorylase - cleavage of 1,4 α glycosidic bond by Pi NOT H2O • Energy of phosphorolysis vs. hydrolysis -low standard state free energy change -transfer potential -driven by Pi concentration -Hydrolysis would require additional step s/ cost of ATP - Think of the difference between adding a phosphate group with hydrolysis • phosphorylation locks glucose in cell (imp. for muscle) • Phosphorylase binds glycogen at storage site and the catalytic site is 4 to 5 glucose residues away from the catalytic site. • Phosphorylase removes 1 residue at a time from glycogen until 4 glucose residues away on either side of 1,6 branch point – stericaly hindered by glycogen storage site • Cleaves without releasing at storage site • general acid/base catalysts • Inorganic phosphate attacks the terminal glucose residue passing through an oxonium ion intermediate. -

(12) Patent Application Publication (10) Pub. No.: US 2009/0292100 A1 Fiene Et Al
US 20090292100A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0292100 A1 Fiene et al. (43) Pub. Date: Nov. 26, 2009 (54) PROCESS FOR PREPARING (86). PCT No.: PCT/EP07/57646 PENTAMETHYLENE 1.5-DIISOCYANATE S371 (c)(1), (75) Inventors: Martin Fiene, Niederkirchen (DE): (2), (4) Date: Jan. 9, 2009 (DE);Eckhard Wolfgang Stroefer, Siegel, Mannheim (30) Foreign ApplicationO O Priority Data Limburgerhof (DE); Stephan Aug. 1, 2006 (EP) .................................. O61182.56.4 Freyer, Neustadt (DE); Oskar Zelder, Speyer (DE); Gerhard Publication Classification Schulz, Bad Duerkheim (DE) (51) Int. Cl. Correspondence Address: CSG 18/00 (2006.01) OBLON, SPIVAK, MCCLELLAND MAIER & CD7C 263/2 (2006.01) NEUSTADT, L.L.P. CI2P I3/00 (2006.01) 194O DUKE STREET CD7C 263/10 (2006.01) ALEXANDRIA, VA 22314 (US) (52) U.S. Cl. ........... 528/85; 560/348; 435/128; 560/347; 560/355 (73) Assignee: BASFSE, LUDWIGSHAFEN (DE) (57) ABSTRACT (21) Appl. No.: 12/373,088 The present invention relates to a process for preparing pen tamethylene 1,5-diisocyanate, to pentamethylene 1,5-diiso (22) PCT Filed: Jul. 25, 2007 cyanate prepared in this way and to the use thereof. US 2009/0292100 A1 Nov. 26, 2009 PROCESS FOR PREPARING ene diisocyanates, especially pentamethylene 1,4-diisocyan PENTAMETHYLENE 1.5-DIISOCYANATE ate. Depending on its preparation, this proportion may be up to several % by weight. 0014. The pentamethylene 1,5-diisocyanate prepared in 0001. The present invention relates to a process for pre accordance with the invention has, in contrast, a proportion of paring pentamethylene 1,5-diisocyanate, to pentamethylene the branched pentamethylene diisocyanate isomers of in each 1.5-diisocyanate prepared in this way and to the use thereof. -

Comparative Transcriptomics Reveals Lineage Specific Evolution of Cold
bioRxiv preprint doi: https://doi.org/10.1101/151431; this version posted June 19, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Evolution of cold response in Pooideae 1 Comparative transcriptomics reveals lineage specific evolution of 2 cold response in Pooideae 3 Lars Grønvold1*, Marian Schubert2*, Simen R. Sandve3, Siri Fjellheim2 and Torgeir R. 4 Hvidsten1,4 5 *Contributed equally 6 1Department of Chemistry, Biotechnology and Food Science, Norwegian University of Life 7 Sciences, NO-1432, Ås, Norway. 8 2Department of Plant Sciences, Norwegian University of Life Sciences, Ås NO-1432, Norway. 9 3Centre for Integrative Genetics (CIGENE), Department of Animal and Aquacultural Sciences, 10 Norwegian University of Life Sciences, NO-1432, Ås, Norway. 11 4Umeå Plant Science Centre, Department of Plant Physiology, Umeå University, SE-90187, 12 Umeå, Sweden. 13 Author for correspondence: 14 Torgeir R. Hvidsten 15 Tel: +4767232491 16 Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/151431; this version posted June 19, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Evolution of cold response in Pooideae 17 Abstract 18 Background: Understanding how complex traits evolve through adaptive changes in gene 19 regulation remains a major challenge in evolutionary biology. -

Molecular Pathogenesis of a New Glycogenosis Caused by a Glycogenin-1 Mutation
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Biochimica et Biophysica Acta 1822 (2012) 493–499 Contents lists available at SciVerse ScienceDirect Biochimica et Biophysica Acta journal homepage: www.elsevier.com/locate/bbadis Molecular pathogenesis of a new glycogenosis caused by a glycogenin-1 mutation Johanna Nilsson a,⁎, Adnan Halim b, Ali-Reza Moslemi a, Anders Pedersen c, Jonas Nilsson b, Göran Larson b, Anders Oldfors a a Department of Pathology, Institute of Biomedicine, University of Gothenburg, Gothenburg, Sweden b Department of Clinical Chemistry and Transfusion Medicine, Institute of Biomedicine, University of Gothenburg, Gothenburg, Sweden c Swedish NMR Centre, Gothenburg, Sweden article info abstract Article history: Glycogenin-1 initiates the glycogen synthesis in skeletal muscle by the autocatalytic formation of a short ol- Received 27 June 2011 igosaccharide at tyrosine 195. Glycogenin-1 catalyzes both the glucose-O-tyrosine linkage and the α1,4 glu- Received in revised form 7 November 2011 cosidic bonds linking the glucose molecules in the oligosaccharide. We recently described a patient with Accepted 28 November 2011 glycogen depletion in skeletal muscle as a result of a non-functional glycogenin-1. The patient carried a Available online 9 December 2011 Thr83Met substitution in glycogenin-1. In this study we have investigated the importance of threonine 83 Keywords: for the catalytic activity of glycogenin-1. Non-glucosylated glycogenin-1 constructs, with various amino Glycogen acid substitutions in position 83 and 195, were expressed in a cell-free expression system and autoglucosy- Glycogenin lated in vitro. -

Conformational Plasticity of Glycogenin and Its Maltosaccharide Substrate During Glycogen Biogenesis
Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis Apirat Chaikuada,1, D. Sean Froesea,1, Georgina Berridgea, Frank von Delfta, Udo Oppermanna,b, and Wyatt W. Yuea,2 aStructural Genomics Consortium, Old Road Research Campus Building, Oxford, United Kingdom OX3 7DQ; and bBotnar Research Centre, National Institute of Health Research Oxford Biomedical Research Unit, Oxford, United Kingdom OX3 7LD Edited by Gregory A. Petsko, Brandeis University, Waltham, MA, and approved October 25, 2011 (received for review August 24, 2011) Glycogenin initiates the synthesis of a maltosaccharide chain cova- to glycogenin catalysis remain evasive. First, substrate-induced lently attached to itself on Tyr195 via a stepwise glucosylation conformational changes to shield the active site, often advocated reaction, priming glycogen synthesis. We have captured crystallo- as integral to the GT catalytic mechanism (5, 10), have not been graphic snapshots of human glycogenin during its reaction cycle, observed for glycogenin to date. In addition, the possibilities of revealing a dynamic conformational switch between ground and intrasubunit (13, 14) and intersubunit (15) glucosylation exist for active states mediated by the sugar donor UDP-glucose. This switch a glycogenin dimer, as the growing maltosaccharide chain at- includes the ordering of a polypeptide stretch containing Tyr195, tached to one subunit may theoretically proceed into the active and major movement of an approximately 30-residue “lid” seg- site of its own or adjacent subunit for further glucosylation. This ment covering the active site. The rearranged lid guides the nas- remains a subject of debate and is complicated by a reported cent maltosaccharide chain into the active site in either an intra- or monomeric form of glycogenin at low micromolar concentration intersubunit mode dependent upon chain length and steric factors (13). -

Genome-Wide Investigation of Cellular Functions for Trna Nucleus
Genome-wide Investigation of Cellular Functions for tRNA Nucleus- Cytoplasm Trafficking in the Yeast Saccharomyces cerevisiae DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Hui-Yi Chu Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2012 Dissertation Committee: Anita K. Hopper, Advisor Stephen Osmani Kurt Fredrick Jane Jackman Copyright by Hui-Yi Chu 2012 Abstract In eukaryotic cells tRNAs are transcribed in the nucleus and exported to the cytoplasm for their essential role in protein synthesis. This export event was thought to be unidirectional. Surprisingly, several lines of evidence showed that mature cytoplasmic tRNAs shuttle between nucleus and cytoplasm and their distribution is nutrient-dependent. This newly discovered tRNA retrograde process is conserved from yeast to vertebrates. Although how exactly the tRNA nuclear-cytoplasmic trafficking is regulated is still under investigation, previous studies identified several transporters involved in tRNA subcellular dynamics. At least three members of the β-importin family function in tRNA nuclear-cytoplasmic intracellular movement: (1) Los1 functions in both the tRNA primary export and re-export processes; (2) Mtr10, directly or indirectly, is responsible for the constitutive retrograde import of cytoplasmic tRNA to the nucleus; (3) Msn5 functions solely in the re-export process. In this thesis I focus on the physiological role(s) of the tRNA nuclear retrograde pathway. One possibility is that nuclear accumulation of cytoplasmic tRNA serves to modulate translation of particular transcripts. To test this hypothesis, I compared expression profiles from non-translating mRNAs and polyribosome-bound translating mRNAs collected from msn5Δ and mtr10Δ mutants and wild-type cells, in fed or acute amino acid starvation conditions.