Chapter Template
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
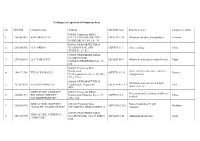
Catalogue of Exporters of Primorsky Krai № ITN/TIN Company Name Address OKVED Code Kind of Activity Country of Export 1 254308
Catalogue of exporters of Primorsky krai № ITN/TIN Company name Address OKVED Code Kind of activity Country of export 690002, Primorsky KRAI, 1 2543082433 KOR GROUP LLC CITY VLADIVOSTOK, PR-T OKVED:51.38 Wholesale of other food products Vietnam OSTRYAKOVA 5G, OF. 94 690001, PRIMORSKY KRAI, 2 2536266550 LLC "SEIKO" VLADIVOSTOK, STR. OKVED:51.7 Other ratailing China TUNGUS, 17, K.1 690003, PRIMORSKY KRAI, VLADIVOSTOK, 3 2531010610 LLC "FORTUNA" OKVED: 46.9 Wholesale trade in specialized stores China STREET UPPERPORTOVA, 38- 101 690003, Primorsky Krai, Vladivostok, Other activities auxiliary related to 4 2540172745 TEK ALVADIS LLC OKVED: 52.29 Panama Verkhneportovaya street, 38, office transportation 301 p-303 p 690088, PRIMORSKY KRAI, Wholesale trade of cars and light 5 2537074970 AVTOTRADING LLC Vladivostok, Zhigura, 46 OKVED: 45.11.1 USA motor vehicles 9KV JOINT-STOCK COMPANY 690091, Primorsky KRAI, Processing and preserving of fish and 6 2504001293 HOLDING COMPANY " Vladivostok, Pologaya Street, 53, OKVED:15.2 China seafood DALMOREPRODUKT " office 308 JOINT-STOCK COMPANY 692760, Primorsky Krai, Non-scheduled air freight 7 2502018358 OKVED:62.20.2 Moldova "AVIALIFT VLADIVOSTOK" CITYARTEM, MKR-N ORBIT, 4 transport 690039, PRIMORSKY KRAI JOINT-STOCK COMPANY 8 2543127290 VLADIVOSTOK, 16A-19 KIROV OKVED:27.42 Aluminum production Japan "ANKUVER" STR. 692760, EDGE OF PRIMORSKY Activities of catering establishments KRAI, for other types of catering JOINT-STOCK COMPANY CITYARTEM, STR. VLADIMIR 9 2502040579 "AEROMAR-ДВ" SAIBEL, 41 OKVED:56.29 China Production of bread and pastry, cakes 690014, Primorsky Krai, and pastries short-term storage JOINT-STOCK COMPANY VLADIVOSTOK, STR. PEOPLE 10 2504001550 "VLADHLEB" AVENUE 29 OKVED:10.71 China JOINT-STOCK COMPANY " MINING- METALLURGICAL 692446, PRIMORSKY KRAI COMPLEX DALNEGORSK AVENUE 50 Mining and processing of lead-zinc 11 2505008358 " DALPOLIMETALL " SUMMER OCTOBER 93 OKVED:07.29.5 ore Republic of Korea 692183, PRIMORSKY KRAI KRAI, KRASNOARMEYSKIY DISTRICT, JOINT-STOCK COMPANY " P. -

China Russia
1 1 1 1 Acheng 3 Lesozavodsk 3 4 4 0 Didao Jixi 5 0 5 Shuangcheng Shangzhi Link? ou ? ? ? ? Hengshan ? 5 SEA OF 5 4 4 Yushu Wuchang OKHOTSK Dehui Mudanjiang Shulan Dalnegorsk Nongan Hailin Jiutai Jishu CHINA Kavalerovo Jilin Jiaohe Changchun RUSSIA Dunhua Uglekamensk HOKKAIDOO Panshi Huadian Tumen Partizansk Sapporo Hunchun Vladivostok Liaoyuan Chaoyang Longjing Yanji Nahodka Meihekou Helong Hunjiang Najin Badaojiang Tong Hua Hyesan Kanggye Aomori Kimchaek AOMORI ? ? 0 AKITA 0 4 DEMOCRATIC PEOPLE'S 4 REPUBLIC OF KOREA Akita Morioka IWATE SEA O F Pyongyang GULF OF KOREA JAPAN Nampo YAMAJGATAA PAN Yamagata MIYAGI Sendai Haeju Niigata Euijeongbu Chuncheon Bucheon Seoul NIIGATA Weonju Incheon Anyang ISIKAWA ChechonREPUBLIC OF HUKUSIMA Suweon KOREA TOTIGI Cheonan Chungju Toyama Cheongju Kanazawa GUNMA IBARAKI TOYAMA PACIFIC OCEAN Nagano Mito Andong Maebashi Daejeon Fukui NAGANO Kunsan Daegu Pohang HUKUI SAITAMA Taegu YAMANASI TOOKYOO YELLOW Ulsan Tottori GIFU Tokyo Matsue Gifu Kofu Chiba SEA TOTTORI Kawasaki KANAGAWA Kwangju Masan KYOOTO Yokohama Pusan SIMANE Nagoya KANAGAWA TIBA ? HYOOGO Kyoto SIGA SIZUOKA ? 5 Suncheon Chinhae 5 3 Otsu AITI 3 OKAYAMA Kobe Nara Shizuoka Yeosu HIROSIMA Okayama Tsu KAGAWA HYOOGO Hiroshima OOSAKA Osaka MIE YAMAGUTI OOSAKA Yamaguchi Takamatsu WAKAYAMA NARA JAPAN Tokushima Wakayama TOKUSIMA Matsuyama National Capital Fukuoka HUKUOKA WAKAYAMA Jeju EHIME Provincial Capital Cheju Oita Kochi SAGA KOOTI City, town EAST CHINA Saga OOITA Major Airport SEA NAGASAKI Kumamoto Roads Nagasaki KUMAMOTO Railroad Lake MIYAZAKI River, lake JAPAN KAGOSIMA Miyazaki International Boundary Provincial Boundary Kagoshima 0 12.5 25 50 75 100 Kilometers Miles 0 10 20 40 60 80 ? ? ? ? 0 5 0 5 3 3 4 4 1 1 1 1 The boundaries and names show n and t he designations us ed on this map do not imply of ficial endors ement or acceptance by the United N at ions. -

Primorsky Krai 165,900 Sq
PRIMORSKY KHABAROVSK Trans- Siberian Railroad Russian Far East Amur River JAO Agzu Samarga Edinka POZHARSKY Svetlaya POZHARSKY Ulunga Verkhne Pereval Vostok Luchegorsk Krasny Yar Kuznetsovo Ignatevka Pozharskoe Maximovka Guberovo Glubinnoe CHINA Amgu !. Dalnerechensk Roshchino TERNEISKY Lazo Novopokrovka KRASNOARMEISKY Rakitnoe Velikaya Kema Tamga DALNERECHENSKY Malaya Kema Melnichnoe !. Lesozavodsk SKY LESOZAVOD Tury Rog Gornye Klyuchi Ternei Ariadnoe Kirovsky ¯ Novokachalinsk Lake lroad Dalny Kut KIROVSKY ORSKY KHANKA Rai Ilinka Plastun Khanka Gorny km DALNEG Cheremshany Dvoryanka berian PO ISKY Samarka Kamen-Rybolov Krasnorechensky 100 GRANICHNY Dalnegorsk !. Zharikovo Tr!.ans-SiSpassk-Dalny KY Koksharovka SPASSKY Kamenka KHOROLS AKOLEVS Khrustalny Pogranichny Khorol Y Yakovlevka EVSKY Rudny Rudnaya Pristan ! CH UGU CHERNIGOVSKY !Kavalerovo EROVSKY KY Lipovtsy ! Sibirtsevo Chuguevka KAVAL ! !. ANUCHINSKY Arsenev Yaroslavsky OKTYABRSKY Vesely Yar n MI Mikhailovka Pokrovka KHAILOVSKY Anuchino Olga a Mikhailovka Arkhipovka Nikolo-Lvovskoe !. OLGINSKY PARTI p USSURIISKY UssuriiskSHKO Terekhovka a ZANSKY TOVSKY Margaritovo NADEZHDINSKY Lazo J Sergeevka Uglovoe Artem ! !. ! Smolyaninovo ZOVSKY LA Valentin f Primorsky !. P! !. Bolshoi Kamen Partizansk o ! Russky Nakhodka Preobrazhenie ! . Popova ! a Kraskino KHASANSKY Poset Slavyanka e VLADIVOSTOK S Map 2.1 Zarubino Khasan Primorsky Krai 165,900 sq. km Newell, J. 2004. The Russian Far East: A Reference Guide for Conservation and Development. McKinleyville, CA: Daniel & Daniel. 466 pages By Newell and Zhou / Sources: Ministry of Natural Resources, 2002; ESRI, 2002. 110 Ⅲ THE RUSSIAN FAR EAST CHAPTER 2 Primorsky Krai PRIMORSKY Location Situated along the southeastern border of the rfe, Primorsky Krai, or Primorie, shares a common border with China in the west and Khabarovsk Krai in the north. To the east lies the Sea of Japan, which separates Primorsky from Japan by only 400 km. -

Subject of the Russian Federation)
How to use the Atlas The Atlas has two map sections The Main Section shows the location of Russia’s intact forest landscapes. The Thematic Section shows their tree species composition in two different ways. The legend is placed at the beginning of each set of maps. If you are looking for an area near a town or village Go to the Index on page 153 and find the alphabetical list of settlements by English name. The Cyrillic name is also given along with the map page number and coordinates (latitude and longitude) where it can be found. Capitals of regions and districts (raiony) are listed along with many other settlements, but only in the vicinity of intact forest landscapes. The reader should not expect to see a city like Moscow listed. Villages that are insufficiently known or very small are not listed and appear on the map only as nameless dots. If you are looking for an administrative region Go to the Index on page 185 and find the list of administrative regions. The numbers refer to the map on the inside back cover. Having found the region on this map, the reader will know which index map to use to search further. If you are looking for the big picture Go to the overview map on page 35. This map shows all of Russia’s Intact Forest Landscapes, along with the borders and Roman numerals of the five index maps. If you are looking for a certain part of Russia Find the appropriate index map. These show the borders of the detailed maps for different parts of the country. -

Phoenix Final Report 2007
Phoenix Fund “Fighting for the Minds II”: strengthening tiger conservation in Primorye, Russian Far East in 2007 Final report January – December 2007 Vladivostok 2007 Phoenix Final Report _____________________________________________________________________________________________ January 01 – December 31, 2007 Contents page I. Project Overview ………………………………………………………………………....3 II. Project Implementation ………………………………………………………………….3 2.1. Education and outreach activities Eco-centre in Partizansk city and environmental education in Partizansky District………………………………………………………………………………………..3 Eco-centre in Luchegorsk city……………………………………………………………..5 Children’s art contests devoted to tigers ………………………………………………...5 Design and publication of calendars with children’s paintings of tiger ……………….6 Journalist Awards …………………………………………………………………………..6 Tiger Day Festival …………………………………………………………………………..6 2.2. Anti-poaching activities Support for Western wildlife managers’ team …………………………………………..9 III. Measurable objectives delivered ……………………………………………………...13 IV. Acknowledgement………………………………………………………………..……...14 V. Attachment…………………………………………………….…………………..……...15 2 Phoenix Final Report _____________________________________________________________________________________________ January 01 – December 31, 2007 I. Project Overview II. Project Implementation Since the year of its establishment in 1998, 2.1. Education and outreach activities the Phoenix Fund has been carrying out nature conservation projects in the south of Eco-centre in Partizansk city and the Russian -

United Nations Code for Trade and Transport Locations (UN/LOCODE) for Russia
United Nations Code for Trade and Transport Locations (UN/LOCODE) for Russia N.B. To check the official, current database of UN/LOCODEs see: https://www.unece.org/cefact/locode/service/location.html UN/LOCODE Location Name State Functionality Status Coordinatesi RU 7RS Shemakha CHE Road terminal; Recognised location 5614N 05915E RU AAD Aleksandrov (Alexandrov) Road terminal; Request under consideration 5623N 03837E RU AAQ Anapa Airport; Code adopted by IATA or ECLAC RU ABA Abakan Road terminal; Recognised location 5342N 09125E RU ABC Ambarchik SA Port; Request under consideration 6937N 16218E RU ABD Abdulino ORE Rail terminal; Road terminal; Recognised location 5342N 05340E RU ABK Abinsk KDA Port; Rail terminal; Road terminal; Recognised location 4452N 03809E RU ABS Akhtubinsk Function not known Recognised location RU ACS Achinsk Airport; Code adopted by IATA or ECLAC RU ADH Aldan Airport; Code adopted by IATA or ECLAC RU ADT Ardatov NIZ Road terminal; Recognised location 5514N 04306E RU AER Sochi KDA Port; Rail terminal; Road terminal; Airport; Code adopted by IATA or ECLAC 4336N 03943E RU AGI Aginskoye Road terminal; QQ RU AGK Angarsk IRK Port; Rail terminal; Road terminal; Recognised location 5232N 10353E RU AHK Arkhangel'skoye STA Road terminal; Recognised location 4436N 04406E RU AHR Akhtari Function not known Request under consideration RU AKS Aksay ROS Port; Request under consideration 4715N 03953E RU ALA Nartkala KB Road terminal; Recognised location 4333N 04351E RU ALE Aleysk AL Rail terminal; Road terminal; Recognised location -

Securing a Future for Amur Leopards and Tigers in Russia
Securing a Future for Amur Leopards and Tigers in Russia – VI 2018 Final Report Phoenix Fund 1 Securing a Future for Amur Leopards and Tigers in Russia – VI • 2018 Final Report TABLE OF CONTENTS Background ................................................................................................................................................... 2 Project Summary ........................................................................................................................................... 3 Project Activities............................................................................................................................................ 4 SMART in five protected areas .................................................................................................................. 4 Annual workshop for educators ................................................................................................................ 8 Education in Khasan, Lazo, Terney and Vladivostok ................................................................................. 9 Tiger Day in Primorye .............................................................................................................................. 11 Art Contest .............................................................................................................................................. 13 Photo credits: PRNCO “Tiger “Centre”, Far Eastern Operational Customs Office, Land of the Leopard National Park, Alexander Ratnikov, and children's paintings -

Structure and Genesis of the Taukha Mesozoic Accretionary Prism (Southern Sikhote-Alin, Russia)
Structure and genesis of the Taukha Mesozoic accretionary prism (southern Sikhote-Alin, Russia) Igor’ V. KEMKIN Far East Geological Institute, Far Eastern Branch, Russian Academy of Sciences, Prospect 100-letiya Vladivostoka, 159, Vladivostok 690022 (Russia) [email protected] Raisa A. KEMKINA Far East State Technical University, Poushkinskaya street, 10, Vladivostok 690090 (Russia) Kemkin I. V. & Kemkina R. A. 2000. — Structure and genesis of the Taukha Mesozoic accretionary prism (southern Sikhote-Alin, Russia). Geodiversitas 22 (4) : 481-491. ABSTRACT Based on a study of several sections of the Taukha terrane in southern Sikhote-Alin comprising lithologic, structural and paleontological analysis, the structure and stratigraphy of the terrane was reconstructed. Three structur- al units are defined for the terrane, each with a discrete origin. Each unit KEY WORDS Terrane, consists of a paleo-oceanic plate formations gradually changing above on a accretionary prism, section by terrigenous rocks, which are further replaced by an olistostrome. accretion, Radiolarian age data for the accretionary prism indicates that fragments of a tectonostratigraphy, radiolaria, late Devonian-Permian, Early Triassic-Late Jurassic and Late Jurassic-Early olistostrome. Cretaceous paleo-oceanic plate were consecutively accreted into the prism. RÉSUMÉ Structure et genèse du prisme d’accrétion mésozoïque de Taukha (Sud Sikhote- Alin, Russie). À partir d’études lithologiques, structurales et paléontologiques, la structure et la stratigraphie du terrain accrété de Taukha (Russie) ont pu être établies. Trois unités structurales ont été définies ; chacune consiste en une série océa- MOTS CLÉS Terranes, nique devenant graduellement terrigène, surmontée par un olistostrome. Les prisme d’accrétion, datations obtenues montrent que ces trois fragments ont été successivement accrétion, accrétés dans un prisme d’accrétion ; les unités ont des âges de plus en plus tectonostratigraphie, radiolaires, jeunes : Dévonien supérieur à Permien, Trias inférieur à Jurassique supérieur olistostrome. -

Annual Report 2008
Annual Report 2008 West Siberian Resources Ltd 2008 Revenues MUSD • Successful merger with Alliance Oil Company Cash fl ow from operations, before changes in working capital MUSD • Cash fl ow increased to MUSD 367 Oil reserve growth Proven and probable reserves • Proven and probable reserves Million barrels increased by 35% Total oil production and refi ning volumes • 24.6 million barrels of oil were Million barrels refi ned and 17.4 million barrels of oil were produced. Crude oil production Oil refi ning Comparative information for 2004–2007 presented in graphs and tables on pages 3–21 refers to data from WSR’s previous dis- closures, including previous annual reports, unless otherwise noted. In the fi nancial statements presented on pages 34–92, all comparative information refers to Alliance Oil Company’s historical fi nancial statements unless otherwise noted. 3 Annual Report 2008 The Annual General Meeting The company’s Annual General Meeting (“AGM”) will be held on May 28, 2009 at 4 pm at the Stockholm Concert Hall (The Grünewald Hall), in Stockholm, Sweden. Holders of Swedish Depository Receipts, (“SDRs”), of the Company who wish to attend the AGM must be listed in the register of directly registered holders of SDRs kept by Eu- roclear Sweden AB on Friday, May 22, 2009 and notify Skandinaviska Enskilda Banken AB (publ) (“SEB”) of their intention to attend the AGM not later than the same day, Friday, May 22, 2009 at 5.00 pm. SDR holders registered in the name of a nominee must have their SDRs re-registered in their own names in the Euroclear Sweden AB register in order to at- tend and vote at the AGM. -

Mitochondrial DNA Variation in Olga Bay Larch (Larix Olgensis A. Henry) from Primorsky Krai of Russia E
ISSN 10227954, Russian Journal of Genetics, 2014, Vol. 50, No. 3, pp. 253–260. © Pleiades Publishing, Inc., 2014. Original Russian Text © E.A. Vasyutkina, G.D. Reunova, A.E. Tupikin, Yu.N. Zhuravlev, 2014, published in Genetika, 2014, Vol. 50, No. 3, pp. 291–298. PLANT GENETICS Mitochondrial DNA Variation in Olga Bay Larch (Larix olgensis A. Henry) from Primorsky Krai of Russia E. A. Vasyutkinaa, G. D. Reunovaa, A. E. Tupikinb, and Yu. N. Zhuravleva aInstitute of Biology and Soil Science, Far Eastern Branch of the Russian Academy of Sciences, Vladivostok, 690022 Russia email: [email protected] bInstitute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, 630090 Russia Received August 8, 2013; in final form, October 10, 2013 Abstract—Two mitochondrial DNA fragments, nad4(3c4r) and nad5(12r), were sequenced in 58 larch accessions from the range of Larix olgensis A. Henry. Combinations of the nad4 polymorphic sites formed four haplotypes, two of which (H3 and H4) were unique and two (H1, H2) were common. Haplotype H1 was found only in pure L. olgensis from the vicinity of Olga Bay and in a number of accessions from the southern part of the range. Haplotype H2 was detected in the other samples from the range of Olga Bay larch, as well as in hybrid forms. Similarly to the nad4(3c4r) fragment, the mtDNA fragment UBC460 was able to differ entiate larch populations from the range of L. olgensis examined. DOI: 10.1134/S1022795414030107 INTRODUCTION species (L. lubarskii, L. komarovii, L. amurensis, L. ochotensis, L. -

Pre-Neogene Tectonics of the Sea-Of-Japan Region: a View from the Russian Side Alexander I Khanchuk
Pre-Neogene tectonics of the Sea-of-Japan region: A view from the Russian side Alexander I Khanchuk 55® 5 Щ (200№9Я) 275-291Я Ш\ Earth Science vol. 55, no. 5, 275—291(2001) Й Ш '? 5 5 # , 275 -— 291 (2001 l£) Earth Science (Chikyu Kagaku) vol. 55, 275-291. 2001 2 7 5 Pre-Neogene tectonics of the Sea-of-Japan region: A view from the Russian side Alexander I Khanchuk* * A b stract This is a review paper presenting an original regional plate-tectonic model based on the data recently obtained on the geological structure and history of the Sea-of-Japan region. Several paleogeographic schemes for the Jurassic-Cretaceous time represent the author’s geodynamic model for the development of this region. This model is clearly distinctive from the previous ones by postulating that the Californian-type transform margin has been one of the most significant geodynamic settings in the Mesozoic-Cenozoic East Asia. The author’s paleogeodynatnic model includes a geologic reconstruction of the pre-Neogene position of the Japanese Islands according to which the Central Sikhote-Alin, Tanakura and Median faults were parts of a single Early Cretaceous tectonic line. In addition, Early Cretaceous and Paleogene transform plate boundaries, Jurassic and Late Cretaceous supra-subduction continental margins, and Early Cretaceous island arc are recognized. The Early Cretaceous strike-slip movements likely resulted in a gigantic S-form structure of Sikhote Alin. Some Early Cretaceous and Paleogene magmatic rocks are recognized to indicate specific geodynamic structures, namely “slab windows”. K e y W o r d s : plate tectonics. -

I. Project Overview
Phoenix Fund Conflict Tiger Cases Resolution in 2004 Programmatic report January 01 – September 30,2004 Vladivostok 2004 Phoenix Programmatic Report _____________________________________________________________________________________________ January 01 – September 30, 2004 PROGRAMMATIC REPORT January 01 – September 30, 2004 Grantor: Save the Tiger Fund Project Name: Conflict Tiger Cases Resolution in 2004 Project #: 2004-0103-027 Grantee: The Phoenix Fund Report Period: January 01 – September 30, 2004 Grant Period: January 01 – December 31, 2004 I. Project overview The goal of the project is to ensure long-term survival of the Siberian tiger and peaceful coexistence of people and tigers in the Russian Far East through timely human-tiger conflicts resolution and outreach activities. II. Project activities For the reported period 36 conflict tiger cases were registered in Primorsky and Khabarovsky krais. It is twice as much of cases in comparison with the same period in 2003. Four of them started at the end of 2003. Two cases were solved by Khabarovsky Hunting Management Department. Anuchinsky district (Primorsky krai) 2 Chernigovsky district (Primorsky krai) 4 Shkotovsky district (Primorsky krai) 3 Dalnerechensky district (Primorsky krai) 2 Krasnoarmeisky district (Primorsky krai) 2 Nadezhdinsky district (Primorsky krai) 3 Chuguevsky district (Primorsky krai) 1 Khasan district (Primorsky krai) 7 Spassky district (Primorsky krai) 1 Kavalerovsky district (Primorsky krai) 2 Ussuriisky district (Primorsky krai) 2 Khabarovsky krai 4 Partizansky district (Primorsky krai) 1 Pozharsky district (Primorsky krai) 1 Kirovsky district (Primorsky krai) 1 © Inspection Tiger In total 36 conflict tiger cases Burning the remains of tiger killed in July 2004, Pozharsky district of Primorye 1. In November 2003 an adult female tigress wandered into a Russian town exhibiting abnormal behavior.