Regulation of Cyclins During the Cell Cycle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cyclin-Dependent Kinases and P53 Pathways Are Activated Independently and Mediate Bax Activation in Neurons After DNA Damage
The Journal of Neuroscience, July 15, 2001, 21(14):5017–5026 Cyclin-Dependent Kinases and P53 Pathways Are Activated Independently and Mediate Bax Activation in Neurons after DNA Damage Erick J. Morris,1 Elizabeth Keramaris,2 Hardy J. Rideout,3 Ruth S. Slack,2 Nicholas J. Dyson,1 Leonidas Stefanis,3 and David S. Park2 1Massachusetts General Hospital Cancer Center, Laboratory of Molecular Oncology, Charlestown, Massachusetts 02129, 2Neuroscience Research Institute, University of Ottawa, Ottawa, Ontario K1H 8M5, Canada, and 3Columbia University, New York, New York 10032 DNA damage has been implicated as one important initiator of ization, and DNA binding that result from DNA damage are not cell death in neuropathological conditions such as stroke. Ac- affected by the inhibition of CDK activity. Conversely, no de- cordingly, it is important to understand the signaling processes crease in retinoblastoma protein (pRb) phosphorylation was that control neuronal death induced by this stimulus. Previous observed in p53-deficient neurons that were treated with camp- evidence has shown that the death of embryonic cortical neu- tothecin. However, either p53 deficiency or the inhibition of rons treated with the DNA-damaging agent camptothecin is CDK activity alone inhibited Bax translocation, cytochrome c dependent on the tumor suppressor p53 and cyclin-dependent release, and caspase-3-like activation. Taken together, our re- kinase (CDK) activity and that the inhibition of either pathway sults indicate that p53 and CDK are activated independently alone leads to enhanced and prolonged survival. We presently and then act in concert to control Bax-mediated apoptosis. show that p53 and CDKs are activated independently on par- allel pathways. -

The Cryoelectron Microscopy Structure of the Human CDK-Activating Kinase
The cryoelectron microscopy structure of the human CDK-activating kinase Basil J. Grebera,b,1,2, Juan M. Perez-Bertoldic, Kif Limd, Anthony T. Iavaronee, Daniel B. Tosoa, and Eva Nogalesa,b,d,f,2 aCalifornia Institute for Quantitative Biosciences (QB3), University of California, Berkeley, CA 94720; bMolecular Biophysics and Integrative Bio-Imaging Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720; cBiophysics Graduate Group, University of California, Berkeley, CA 94720; dDepartment of Molecular and Cell Biology, University of California, Berkeley, CA 94720; eQB3/Chemistry Mass Spectrometry Facility, University of California, Berkeley, CA 94720; and fHoward Hughes Medical Institute, University of California, Berkeley, CA 94720 Edited by Seth A. Darst, Rockefeller University, New York, NY, and approved August 4, 2020 (received for review May 14, 2020) The human CDK-activating kinase (CAK), a complex composed of phosphoryl transfer (11). However, in addition to cyclin binding, cyclin-dependent kinase (CDK) 7, cyclin H, and MAT1, is a critical full activation of cell cycle CDKs requires phosphorylation of the regulator of transcription initiation and the cell cycle. It acts by T-loop (9, 12). In animal cells, these activating phosphorylations phosphorylating the C-terminal heptapeptide repeat domain of are carried out by CDK7 (13, 14), itself a cyclin-dependent ki- the RNA polymerase II (Pol II) subunit RPB1, which is an important nase whose activity depends on cyclin H (14). regulatory event in transcription initiation by Pol II, and it phos- In human and other metazoan cells, regulation of transcription phorylates the regulatory T-loop of CDKs that control cell cycle initiation by phosphorylation of the Pol II-CTD and phosphor- progression. -

The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression
International Journal of Molecular Sciences Review The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression Tingting Zou and Zhenghong Lin * School of Life Sciences, Chongqing University, Chongqing 401331, China; [email protected] * Correspondence: [email protected] Abstract: The cell cycle is a collection of events by which cellular components such as genetic materials and cytoplasmic components are accurately divided into two daughter cells. The cell cycle transition is primarily driven by the activation of cyclin-dependent kinases (CDKs), which activities are regulated by the ubiquitin-mediated proteolysis of key regulators such as cyclins, CDK inhibitors (CKIs), other kinases and phosphatases. Thus, the ubiquitin-proteasome system (UPS) plays a pivotal role in the regulation of the cell cycle progression via recognition, interaction, and ubiquitination or deubiquitination of key proteins. The illegitimate degradation of tumor suppressor or abnormally high accumulation of oncoproteins often results in deregulation of cell proliferation, genomic instability, and cancer occurrence. In this review, we demonstrate the diversity and complexity of the regulation of UPS machinery of the cell cycle. A profound understanding of the ubiquitination machinery will provide new insights into the regulation of the cell cycle transition, cancer treatment, and the development of anti-cancer drugs. Keywords: cell cycle regulation; CDKs; cyclins; CKIs; UPS; E3 ubiquitin ligases; Deubiquitinases (DUBs) Citation: Zou, T.; Lin, Z. The Involvement of Ubiquitination Machinery in Cell Cycle Regulation and Cancer Progression. 1. Introduction Int. J. Mol. Sci. 2021, 22, 5754. https://doi.org/10.3390/ijms22115754 The cell cycle is a ubiquitous, complex, and highly regulated process that is involved in the sequential events during which a cell duplicates its genetic materials, grows, and di- Academic Editors: Kwang-Hyun Bae vides into two daughter cells. -

A Haploid Genetic Screen Identifies the G1/S Regulatory Machinery As a Determinant of Wee1 Inhibitor Sensitivity
A haploid genetic screen identifies the G1/S regulatory machinery as a determinant of Wee1 inhibitor sensitivity Anne Margriet Heijinka, Vincent A. Blomenb, Xavier Bisteauc, Fabian Degenera, Felipe Yu Matsushitaa, Philipp Kaldisc,d, Floris Foijere, and Marcel A. T. M. van Vugta,1 aDepartment of Medical Oncology, University Medical Center Groningen, University of Groningen, 9723 GZ Groningen, The Netherlands; bDivision of Biochemistry, The Netherlands Cancer Institute, 1066 CX Amsterdam, The Netherlands; cInstitute of Molecular and Cell Biology, Agency for Science, Technology and Research, Proteos#3-09, Singapore 138673, Republic of Singapore; dDepartment of Biochemistry, National University of Singapore, Singapore 117597, Republic of Singapore; and eEuropean Research Institute for the Biology of Ageing, University of Groningen, University Medical Center Groningen, 9713 AV Groningen, The Netherlands Edited by Stephen J. Elledge, Harvard Medical School, Boston, MA, and approved October 21, 2015 (received for review March 17, 2015) The Wee1 cell cycle checkpoint kinase prevents premature mitotic Wee1 kinase at tyrosine (Tyr)-15 to prevent unscheduled Cdk1 entry by inhibiting cyclin-dependent kinases. Chemical inhibitors activity (5, 6). Conversely, timely activation of Cdk1 depends on of Wee1 are currently being tested clinically as targeted anticancer Tyr-15 dephosphorylation by one of the Cdc25 phosphatases drugs. Wee1 inhibition is thought to be preferentially cytotoxic in (7–10). When DNA is damaged, the downstream DNA damage p53-defective cancer cells. However, TP53 mutant cancers do not response (DDR) kinases Chk1 and Chk2 inhibit Cdc25 phos- respond consistently to Wee1 inhibitor treatment, indicating the phatases through direct phosphorylation, which blocks Cdk1 existence of genetic determinants of Wee1 inhibitor sensitivity other activation (11–13). -

Targeting Cyclin-Dependent Kinase 9 and Myeloid Cell Leukaemia 1 in MYC-Driven B-Cell Lymphoma
Targeting cyclin-dependent kinase 9 and myeloid cell leukaemia 1 in MYC-driven B-cell lymphoma Gareth Peter Gregory ORCID ID: 0000-0002-4170-0682 Thesis for Doctor of Philosophy September 2016 Sir Peter MacCallum Department of Oncology The University of Melbourne Doctor of Philosophy Submitted in total fulfilment of the degree of Abstract Aggressive B-cell lymphomas include diffuse large B-cell lymphoma, Burkitt lymphoma and intermediate forms. Despite high response rates to conventional immuno-chemotherapeutic approaches, an unmet need for novel therapeutic by resistance to chemotherapy and radiotherapy. The proto-oncogene MYC is strategies is required in the setting of relapsed and refractory disease, typified frequently dysregulated in the aggressive B-cell lymphomas, however, it has proven an elusive direct therapeutic target. MYC-dysregulated disease maintains a ‘transcriptionally-addicted’ state, whereby perturbation of A significant body of evidence is accumulating to suggest that RNA polymerase II activity may indirectly antagonise MYC activity. Furthermore, very recent studies implicate anti-apoptotic myeloid cell leukaemia 1 (MCL-1) as a critical survival determinant of MYC-driven lymphoma. This thesis utilises pharmacologic and genetic techniques in MYC-driven models of aggressive B-cell lymphoma to demonstrate that cyclin-dependent kinase 9 (CDK9) and MCL-1 are oncogenic dependencies of this subset of disease. The cyclin-dependent kinase inhibitor, dinaciclib, and more selective CDK9 inhibitors downregulation of MCL1 are used -

Datasheet for CDK1-Cyclin B
of T161 is required for activation of the CDK1- Supplied in: 100 mM NaCl, 50 mM HEPES Specific Activity: ~1,000,000 units/mg cyclin B complex and is mediated by the CDK (pH 7.5 @ 25°C), 0.1 mM EDTA, 1 mM DTT, 0.01% CDK1-cyclin B activating kinase (CAK). During G2 phase, Brij 35 and 50% glycerol. Molecular Weight: CDK1 (34 kDa), cyclin B CDK1-cyclin B complex is held in an inactive state (48 kDa). The apparent molecular weight of cyclin by phosphorylation of CDK1 at the two negative Reagents Supplied with Enzyme: B on SDS-PAGE is about 60 kDa. 1-800-632-7799 regulatory sites, T14 and Y15 by CDK1 inhibitory 10X NEBuffer for Protein Kinases (PK). [email protected] protein kinases, Myt1 and Wee1 respectively. Quality Control Assays www.neb.com Dephosphorylation of T14 and Y15 by cell division Reaction Conditions: 1X NEBuffer for PK (NEB Protease Activity: After incubation of 100 units #B6022), supplemented with 200 µM ATP (NEB P6020S 032120413041 cycle 25 (CDC25) protein phosphatase in late G2 of CDK1-cyclin B with a standard mixture phase activates the CDK1-cyclin B complex and #P0756) and gamma-labeled ATP to a final specific of proteins for 2 hours at 30°C, no proteolytic activity of 100–500 µCi/µmol. Incubate at 30°C. triggers the initiation of mitosis. During expression activity could be detected by SDS-PAGE analysis. P6020S in insect cells, the recombiniant CDK1-cyclin B is 1X NEBuffer for PK: Phosphatase Activity: After incubation of 200 units 20,000 U/ml Lot: 0321204 activated in vivo by endogenous kinase (1–4). -

Cyclin a Triggers Mitosis Either Via Greatwall Or Cyclin B
bioRxiv preprint doi: https://doi.org/10.1101/501684; this version posted December 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Cyclin A triggers Mitosis either via Greatwall or Cyclin B Nadia Hégarat(1)*, Adrijana Crncec(1)*, Maria F. Suarez Peredoa Rodri- guez(1), Fabio Echegaray Iturra(1), Yan Gu(1), Paul F. Lang(2), Alexis R. Barr(3), Chris Bakal(4), Masato T. Kanemaki(5), Angus I. Lamond(6), Bela Novak(2), Tony Ly(7)•• and Helfrid Hochegger(1)•• (1) Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Brighton BN19RQ, UK (2) Department of Biochemistry, University of Oxford, South Park Road, Oxford OX13QU, UK (3) MRC London Institute of Medical Science, Imperial College, London W12 0NN, UK (4) The Institute of Cancer Research, London SW3 6JB, UK (5) National Institute of Genetics, Research Organization of Information and Sys- tems (ROIS), and Department of Genetics, SOKENDAI (The Graduate University of Advanced Studies), Yata 1111, Mishima, Shizuoka 411-8540, Japan. (6) Centre for Gene Regulation and Expression, School of Life Sciences, University of Dundee, Dundee DD1 5EH, UK (7) Wellcome Trust Centre for Cell Biology, University of Edinburgh, Edinburgh EH9 3BF, UK * Equal contribution ** Correspondence: Tony Ly: [email protected]; Helfrid Hochegger: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/501684; this version posted December 20, 2018. -
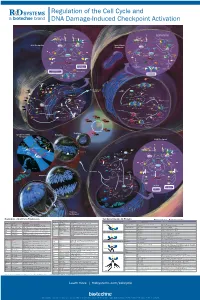
Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation
RnDSy-lu-2945 Regulation of the Cell Cycle and DNA Damage-Induced Checkpoint Activation IR UV IR Stalled Replication Forks/ BRCA1 Rad50 Long Stretches of ss-DNA Rad50 Mre11 BRCA1 Nbs1 Rad9-Rad1-Hus1 Mre11 RPA MDC1 γ-H2AX DNA Pol α/Primase RFC2-5 MDC1 Nbs1 53BP1 MCM2-7 53BP1 γ-H2AX Rad17 Claspin MCM10 Rad9-Rad1-Hus1 TopBP1 CDC45 G1/S Checkpoint Intra-S-Phase RFC2-5 ATM ATR TopBP1 Rad17 ATRIP ATM Checkpoint Claspin Chk2 Chk1 Chk2 Chk1 ATR Rad50 ATRIP Mre11 FANCD2 Ubiquitin MDM2 MDM2 Nbs1 CDC25A Rad50 Mre11 BRCA1 Ub-mediated Phosphatase p53 CDC25A Ubiquitin p53 FANCD2 Phosphatase Degradation Nbs1 p53 p53 CDK2 p21 p21 BRCA1 Ub-mediated SMC1 Degradation Cyclin E/A SMC1 CDK2 Slow S Phase CDC45 Progression p21 DNA Pol α/Primase Slow S Phase p21 Cyclin E Progression Maintenance of Inhibition of New CDC6 CDT1 CDC45 G1/S Arrest Origin Firing ORC MCM2-7 MCM2-7 Recovery of Stalled Replication Forks Inhibition of MCM10 MCM10 Replication Origin Firing DNA Pol α/Primase ORI CDC6 CDT1 MCM2-7 ORC S Phase Delay MCM2-7 MCM10 MCM10 ORI Geminin EGF EGF R GAB-1 CDC6 CDT1 ORC MCM2-7 PI 3-Kinase p70 S6K MCM2-7 S6 Protein Translation Pre-RC (G1) GAB-2 MCM10 GSK-3 TSC1/2 MCM10 ORI PIP2 TOR Promotes Replication CAK EGF Origin Firing Origin PIP3 Activation CDK2 EGF R Akt CDC25A PDK-1 Phosphatase Cyclin E/A SHIP CIP/KIP (p21, p27, p57) (Active) PLCγ PP2A (Active) PTEN CDC45 PIP2 CAK Unwinding RPA CDC7 CDK2 IP3 DAG (Active) Positive DBF4 α Feedback CDC25A DNA Pol /Primase Cyclin E Loop Phosphatase PKC ORC RAS CDK4/6 CDK2 (Active) Cyclin E MCM10 CDC45 RPA IP Receptor -

Human P14arf-Mediated Cell Cycle Arrest Strictly Depends on Intact P53 Signaling Pathways
Oncogene (2002) 21, 3207 ± 3212 ã 2002 Nature Publishing Group All rights reserved 0950 ± 9232/02 $25.00 www.nature.com/onc Human p14ARF-mediated cell cycle arrest strictly depends on intact p53 signaling pathways H Oliver Weber1,2, Temesgen Samuel1,3, Pia Rauch1 and Jens Oliver Funk*,1 1Laboratory of Molecular Tumor Biology, Department of Dermatology, University of Erlangen-Nuremberg, 91052 Erlangen, Germany; 2Regulation of Cell Growth Laboratory, National Cancer Institute, Frederick, Maryland, MD 21702-1201, USA The tumor suppressor ARF is transcribed from the INK4a/ 4 and 6 activities, thus leading to decreased phosphor- ARF locus in partly overlapping reading frames with the ylation of RB and to G1 arrest. Cells that are de®cient CDK inhibitor p16Ink4a. ARF is able to antagonize the for RB are resistant to p16Ink4a-mediated cell cycle MDM2-mediated ubiquitination and degradation of p53, arrest (Sherr and Roberts, 1995; Weinberg, 1995). leading to either cell cycle arrest or apoptosis, depending on ARF is also a cell cyle inhibitor. It does not directly the cellular context. However, recent data point to inhibit CDKs but interferes with the function of additional p53-independent functions of mouse p19ARF. MDM2 to destabilize p53. ARF may be activated by Little is known about the dependency of human p14ARF aberrant activation of oncoproteins such as Ras function on p53 and its downstream genes. Therefore, we (Palmero et al., 1998), c-myc (Zindy et al., 1998), analysed the mechanism of p14ARF-induced cell cycle arrest E1A (de Stanchina et al., 1998), Abl (Radfar et al., in several human cell types. -

21-Containing C.Yclin Kinases Exist in Oth Acnve and Lnacnve States
Downloaded from genesdev.cshlp.org on October 5, 2021 - Published by Cold Spring Harbor Laboratory Press 21-containing c.yclin kinases exist in oth acnve and lnacnve states Hui Zhang, Gregory J. Hannon, and David Beach Howard Hughes Medical Institute, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724 USA In normal fibroblasts CDKs exist predominantly in p21/PCNA/cyclin/CDK quaternary complexes, whereas in p53-deficient cells, p21 expression is depressed and the kinases are reduced to a cyclin/CDK binary state, p21 is a universal cyclin kinase inhibitor, but we show here that p21-containing complexes exist in both catalytically active and inactive forms. This finding challenges the current view that active cyclin kinases function only in the binary state and reveals the subtlety with which tumor-suppressor proteins modulate the cell cycle. [Key Words: p21-containing cyclin kinase; cell cycle progression; human fibroblasts; quaternary complex; kinase inhibitor] Received May 12, 1994; revised version accepted June 16, 1994. Much of our current understanding of the regulation of their relative affinities vary with each enzyme (Gu et al. the cell division cycle has emerged from studies of a 1993; Harper et al. 1993; Xiong et al. 1993b). Further- family of protein kinases [cdc2, CDC28, and generically more, several lines of evidence suggest that p21 expres- cyclin-dependent kinase (CDK)] and their inhibitors and sion is regulated by the p53 tumor-suppressor protein activators (for review, see Sherr 1993). A critical step in (E1-Deiry et al. 1993; Xiong et al. 1993b). Thus, cells understanding cell cycle control was the discovery that derived from certain p53-deficient Li-Fraumeni patients CDKs interact with cyclins, proteins that serve as essen- lack p21 associated with cyclin kinases (Xiong et al. -

NF-Y Binds to Both G1- and G2-Specific Cyclin Promoters; a Possible Role in Linking CDK2/Cyclin a to CDK1/Cyclin B
BMB reports NF-Y binds to both G1- and G2-specific cyclin promoters; a possible role in linking CDK2/Cyclin A to CDK1/Cyclin B Hee-Don Chae#, Jungbin Kim# & Deug Y. Shin* Department of Microbiology, Dankook University College of Medicine, Cheonan, 330-714, Korea We previously reported that CDK2/Cyclin A can phosphor- standing of cell growth and division. ylate and activate the transcription factor NF-Y. In this study, It is well established that retinoblastoma (Rb) tumor sup- we investigated a potential regulatory role for NF-Y in the tran- pressor proteins coordinate the sequential activation of CDK4 scription of Cyclin A and other cell cycle regulatory genes. and CDK2 during the G1/S transition (3, 4). The CDK4/Cyclin Gel-shift assays demonstrate that NF-Y binds to CCAAT se- D complex phosphorylates Rb, promoting Cyclin E expression quences in the Cyclin A promoter, as well as to those in the through its E2F-dependent transcriptional activation and dis- promoters of cell cycle G2 regulators such as CDC2, Cyclin B ruption of the Rb-HDAC complex, culminating in CDK2 kin- and CDC25C. Furthermore, expression of Cyclin A increases ase activation as S phase progresses (5, 6). In contrast, the fac- NF-Y’s affinity for CCAAT sequences in the CDC2 promoter; tors involved in the handoff from CDK2 to CDK1 remain however, Cyclin A’s induction of CDC2 transcription is antag- unidentified. However, previous work by our group and others onized by p21, an inhibitor of CDK2/Cyclin A. These results demonstrates that overexpression of p53 blocks the G2/M tran- suggest a model wherein NF-Y binds to and activates tran- sition (7, 8) and inhibits cell cycle-dependent transcription of scription from the Cyclin A promoter, increasing cellular levels CDC2 and CCNB genes with consequent CDK1 inactivation of Cyclin A/CDK2 and potentiating NF-Y’s capacity for tran- (7). -

Cytometry of Cyclin Proteins
Reprinted with permission of Cytometry Part A, John Wiley and Sons, Inc. Cytometry of Cyclin Proteins Zbigniew Darzynkiewicz, Jianping Gong, Gloria Juan, Barbara Ardelt, and Frank Traganos The Cancer Research Institute, New York Medical College, Valhalla, New York Received for publication January 22, 1996; accepted March 11, 1996 Cyclins are key components of the cell cycle pro- gests that the partner kinase CDK4 (which upon ac- gression machinery. They activate their partner cy- tivation by D-type cyclins phosphorylates pRB com- clin-dependent kinases (CDKs) and possibly target mitting the cell to enter S) is perpetually active them to respective substrate proteins within the throughout the cell cycle in these tumor lines. Ex- cell. CDK-mediated phosphorylation of specsc sets pression of cyclin D also may serve to discriminate of proteins drives the cell through particular phases Go vs. GI cells and, as an activation marker, to iden- or checkpoints of the cell cycle. During unper- tify the mitogenically stimulated cells entering the turbed growth of normal cells, the timing of expres- cell cycle. Differences in cyclin expression make it sion of several cyclins is discontinuous, occurring possible to discrirmna* te between cells having the at discrete and well-defined periods of the cell cy- same DNA content but residing at different phases cle. Immunocytochemical detection of cyclins in such as in G2vs. M or G,/M of a lower DNA ploidy vs. relation to cell cycle position (DNA content) by GI cells of a higher ploidy. The expression of cyclins multiparameter flow cytometry has provided a new D, E, A and B1 provides new cell cycle landmarks approach to cell cycle studies.