Downloaded the Normalized Gene Expression Profile from the GEO (Package Geoquery (5))
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Mouse Steap4 Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Steap4 Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Steap4 conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Steap4 gene (NCBI Reference Sequence: NM_054098 ; Ensembl: ENSMUSG00000012428 ) is located on Mouse chromosome 5. 5 exons are identified, with the ATG start codon in exon 2 and the TAA stop codon in exon 5 (Transcript: ENSMUST00000115421). Exon 2~3 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Steap4 gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP23-212F6 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for a knock-out allele exhibit adipose accumulation, oxidative stress, increased liver weight, lower metabolic rate, hypoactivity, insulin resistance, glucose intolerance, mild hyperglycemia and dyslipidemia. Exon 2 starts from about 100% of the coding region. The knockout of Exon 2~3 will result in frameshift of the gene. The size of intron 1 for 5'-loxP site insertion: 14897 bp, and the size of intron 3 for 3'-loxP site insertion: 1385 bp. The size of effective cKO region: ~2082 bp. The cKO region does not have any other known gene. Page 1 of 7 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele 5' gRNA region gRNA region 3' 1 2 3 4 5 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Exon of mouse Steap4 Homology arm cKO region loxP site Page 2 of 7 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Analysis of the Indacaterol-Regulated Transcriptome in Human Airway
Supplemental material to this article can be found at: http://jpet.aspetjournals.org/content/suppl/2018/04/13/jpet.118.249292.DC1 1521-0103/366/1/220–236$35.00 https://doi.org/10.1124/jpet.118.249292 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 366:220–236, July 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Analysis of the Indacaterol-Regulated Transcriptome in Human Airway Epithelial Cells Implicates Gene Expression Changes in the s Adverse and Therapeutic Effects of b2-Adrenoceptor Agonists Dong Yan, Omar Hamed, Taruna Joshi,1 Mahmoud M. Mostafa, Kyla C. Jamieson, Radhika Joshi, Robert Newton, and Mark A. Giembycz Departments of Physiology and Pharmacology (D.Y., O.H., T.J., K.C.J., R.J., M.A.G.) and Cell Biology and Anatomy (M.M.M., R.N.), Snyder Institute for Chronic Diseases, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada Received March 22, 2018; accepted April 11, 2018 Downloaded from ABSTRACT The contribution of gene expression changes to the adverse and activity, and positive regulation of neutrophil chemotaxis. The therapeutic effects of b2-adrenoceptor agonists in asthma was general enriched GO term extracellular space was also associ- investigated using human airway epithelial cells as a therapeu- ated with indacaterol-induced genes, and many of those, in- tically relevant target. Operational model-fitting established that cluding CRISPLD2, DMBT1, GAS1, and SOCS3, have putative jpet.aspetjournals.org the long-acting b2-adrenoceptor agonists (LABA) indacaterol, anti-inflammatory, antibacterial, and/or antiviral activity. Numer- salmeterol, formoterol, and picumeterol were full agonists on ous indacaterol-regulated genes were also induced or repressed BEAS-2B cells transfected with a cAMP-response element in BEAS-2B cells and human primary bronchial epithelial cells by reporter but differed in efficacy (indacaterol $ formoterol . -

Human Lectins, Their Carbohydrate Affinities and Where to Find Them
biomolecules Review Human Lectins, Their Carbohydrate Affinities and Where to Review HumanFind Them Lectins, Their Carbohydrate Affinities and Where to FindCláudia ThemD. Raposo 1,*, André B. Canelas 2 and M. Teresa Barros 1 1, 2 1 Cláudia D. Raposo * , Andr1 é LAQVB. Canelas‐Requimte,and Department M. Teresa of Chemistry, Barros NOVA School of Science and Technology, Universidade NOVA de Lisboa, 2829‐516 Caparica, Portugal; [email protected] 12 GlanbiaLAQV-Requimte,‐AgriChemWhey, Department Lisheen of Chemistry, Mine, Killoran, NOVA Moyne, School E41 of ScienceR622 Co. and Tipperary, Technology, Ireland; canelas‐ [email protected] NOVA de Lisboa, 2829-516 Caparica, Portugal; [email protected] 2* Correspondence:Glanbia-AgriChemWhey, [email protected]; Lisheen Mine, Tel.: Killoran, +351‐212948550 Moyne, E41 R622 Tipperary, Ireland; [email protected] * Correspondence: [email protected]; Tel.: +351-212948550 Abstract: Lectins are a class of proteins responsible for several biological roles such as cell‐cell in‐ Abstract:teractions,Lectins signaling are pathways, a class of and proteins several responsible innate immune for several responses biological against roles pathogens. such as Since cell-cell lec‐ interactions,tins are able signalingto bind to pathways, carbohydrates, and several they can innate be a immuneviable target responses for targeted against drug pathogens. delivery Since sys‐ lectinstems. In are fact, able several to bind lectins to carbohydrates, were approved they by canFood be and a viable Drug targetAdministration for targeted for drugthat purpose. delivery systems.Information In fact, about several specific lectins carbohydrate were approved recognition by Food by andlectin Drug receptors Administration was gathered for that herein, purpose. plus Informationthe specific organs about specific where those carbohydrate lectins can recognition be found by within lectin the receptors human was body. -

Single-Cell Analysis Uncovers Fibroblast Heterogeneity
ARTICLE https://doi.org/10.1038/s41467-020-17740-1 OPEN Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination ✉ Lars Muhl 1,2 , Guillem Genové 1,2, Stefanos Leptidis 1,2, Jianping Liu 1,2, Liqun He3,4, Giuseppe Mocci1,2, Ying Sun4, Sonja Gustafsson1,2, Byambajav Buyandelger1,2, Indira V. Chivukula1,2, Åsa Segerstolpe1,2,5, Elisabeth Raschperger1,2, Emil M. Hansson1,2, Johan L. M. Björkegren 1,2,6, Xiao-Rong Peng7, ✉ Michael Vanlandewijck1,2,4, Urban Lendahl1,8 & Christer Betsholtz 1,2,4 1234567890():,; Many important cell types in adult vertebrates have a mesenchymal origin, including fibro- blasts and vascular mural cells. Although their biological importance is undisputed, the level of mesenchymal cell heterogeneity within and between organs, while appreciated, has not been analyzed in detail. Here, we compare single-cell transcriptional profiles of fibroblasts and vascular mural cells across four murine muscular organs: heart, skeletal muscle, intestine and bladder. We reveal gene expression signatures that demarcate fibroblasts from mural cells and provide molecular signatures for cell subtype identification. We observe striking inter- and intra-organ heterogeneity amongst the fibroblasts, primarily reflecting differences in the expression of extracellular matrix components. Fibroblast subtypes localize to discrete anatomical positions offering novel predictions about physiological function(s) and regulatory signaling circuits. Our data shed new light on the diversity of poorly defined classes of cells and provide a foundation for improved understanding of their roles in physiological and pathological processes. 1 Karolinska Institutet/AstraZeneca Integrated Cardio Metabolic Centre, Blickagången 6, SE-14157 Huddinge, Sweden. -
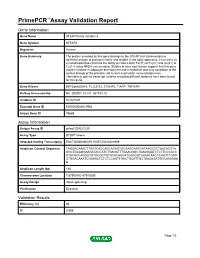
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name STEAP family member 4 Gene Symbol STEAP4 Organism Human Gene Summary The protein encoded by this gene belongs to the STEAP (six transmembrane epithelial antigen of prostate) family and resides in the golgi apparatus. It functions as a metalloreductase that has the ability to reduce both Fe(3+) to Fe(2+) and Cu(2+) to Cu(1+) using NAD(+) as acceptor. Studies in mice and human suggest that this gene maybe involved in adipocyte development and metabolism and may contribute to the normal biology of the prostate cell as well as prostate cancer progression. Alternatively spliced transcript variants encoding different isoforms have been found for this gene. Gene Aliases DKFZp666D049, FLJ23153, STAMP2, TIARP, TNFAIP9 RefSeq Accession No. NC_000007.13, NT_007933.15 UniGene ID Hs.521008 Ensembl Gene ID ENSG00000127954 Entrez Gene ID 79689 Assay Information Unique Assay ID qHsaCID0021230 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000380079, ENST00000301959 Amplicon Context Sequence TAGGACAAACTTGATCACCAGCACAGTGCAAGGAATGATAAGCCCTAACACGTA GGCTGCAGGAAGATACCATCTGAGATTTGAAGGGCTGAGGAATCTCTTCCCACC GTACACCAGGGTGTGGGCTGTACACAAGATCAGGGTCAAATAACCCAGTTTGGA CTGGACAAATCGGAACTCTCTCCAGTTGACTGCATTGCTAACAGATGGCAAAGAA G Amplicon Length (bp) 188 Chromosome Location 7:87908782-87910285 Assay Design Intron-spanning Purification Desalted Validation Results Efficiency (%) 96 R2 0.999 Page 1/5 PrimePCR™Assay Validation Report cDNA Cq 22.3 cDNA Tm (Celsius) 84 gDNA Cq 38.74 Specificity (%) 100 Information -

ID AKI Vs Control Fold Change P Value Symbol Entrez Gene Name *In
ID AKI vs control P value Symbol Entrez Gene Name *In case of multiple probesets per gene, one with the highest fold change was selected. Fold Change 208083_s_at 7.88 0.000932 ITGB6 integrin, beta 6 202376_at 6.12 0.000518 SERPINA3 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 1553575_at 5.62 0.0033 MT-ND6 NADH dehydrogenase, subunit 6 (complex I) 212768_s_at 5.50 0.000896 OLFM4 olfactomedin 4 206157_at 5.26 0.00177 PTX3 pentraxin 3, long 212531_at 4.26 0.00405 LCN2 lipocalin 2 215646_s_at 4.13 0.00408 VCAN versican 202018_s_at 4.12 0.0318 LTF lactotransferrin 203021_at 4.05 0.0129 SLPI secretory leukocyte peptidase inhibitor 222486_s_at 4.03 0.000329 ADAMTS1 ADAM metallopeptidase with thrombospondin type 1 motif, 1 1552439_s_at 3.82 0.000714 MEGF11 multiple EGF-like-domains 11 210602_s_at 3.74 0.000408 CDH6 cadherin 6, type 2, K-cadherin (fetal kidney) 229947_at 3.62 0.00843 PI15 peptidase inhibitor 15 204006_s_at 3.39 0.00241 FCGR3A Fc fragment of IgG, low affinity IIIa, receptor (CD16a) 202238_s_at 3.29 0.00492 NNMT nicotinamide N-methyltransferase 202917_s_at 3.20 0.00369 S100A8 S100 calcium binding protein A8 215223_s_at 3.17 0.000516 SOD2 superoxide dismutase 2, mitochondrial 204627_s_at 3.04 0.00619 ITGB3 integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) 223217_s_at 2.99 0.00397 NFKBIZ nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta 231067_s_at 2.97 0.00681 AKAP12 A kinase (PRKA) anchor protein 12 224917_at 2.94 0.00256 VMP1/ mir-21likely ortholog -

Supplementary Table S1. List of Differentially Expressed
Supplementary table S1. List of differentially expressed transcripts (FDR adjusted p‐value < 0.05 and −1.4 ≤ FC ≥1.4). 1 ID Symbol Entrez Gene Name Adj. p‐Value Log2 FC 214895_s_at ADAM10 ADAM metallopeptidase domain 10 3,11E‐05 −1,400 205997_at ADAM28 ADAM metallopeptidase domain 28 6,57E‐05 −1,400 220606_s_at ADPRM ADP‐ribose/CDP‐alcohol diphosphatase, manganese dependent 6,50E‐06 −1,430 217410_at AGRN agrin 2,34E‐10 1,420 212980_at AHSA2P activator of HSP90 ATPase homolog 2, pseudogene 6,44E‐06 −1,920 219672_at AHSP alpha hemoglobin stabilizing protein 7,27E‐05 2,330 aminoacyl tRNA synthetase complex interacting multifunctional 202541_at AIMP1 4,91E‐06 −1,830 protein 1 210269_s_at AKAP17A A‐kinase anchoring protein 17A 2,64E‐10 −1,560 211560_s_at ALAS2 5ʹ‐aminolevulinate synthase 2 4,28E‐06 3,560 212224_at ALDH1A1 aldehyde dehydrogenase 1 family member A1 8,93E‐04 −1,400 205583_s_at ALG13 ALG13 UDP‐N‐acetylglucosaminyltransferase subunit 9,50E‐07 −1,430 207206_s_at ALOX12 arachidonate 12‐lipoxygenase, 12S type 4,76E‐05 1,630 AMY1C (includes 208498_s_at amylase alpha 1C 3,83E‐05 −1,700 others) 201043_s_at ANP32A acidic nuclear phosphoprotein 32 family member A 5,61E‐09 −1,760 202888_s_at ANPEP alanyl aminopeptidase, membrane 7,40E‐04 −1,600 221013_s_at APOL2 apolipoprotein L2 6,57E‐11 1,600 219094_at ARMC8 armadillo repeat containing 8 3,47E‐08 −1,710 207798_s_at ATXN2L ataxin 2 like 2,16E‐07 −1,410 215990_s_at BCL6 BCL6 transcription repressor 1,74E‐07 −1,700 200776_s_at BZW1 basic leucine zipper and W2 domains 1 1,09E‐06 −1,570 222309_at -

Single-Cell RNA Sequencing of Human, Macaque, and Mouse Testes Uncovers Conserved and Divergent Features of Mammalian Spermatogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.03.17.994509; this version posted March 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Single-cell RNA sequencing of human, macaque, and mouse testes uncovers conserved and divergent features of mammalian spermatogenesis Adrienne Niederriter Shami1,7, Xianing Zheng1,7, Sarah K. Munyoki2,7, Qianyi Ma1, Gabriel L. Manske3, Christopher D. Green1, Meena Sukhwani2, , Kyle E. Orwig2*, Jun Z. Li1,4*, Saher Sue Hammoud1,3,5,6,8* 1Department of Human Genetics, University of Michigan, Ann Arbor, MI, USA 2 Department of Obstetrics, Gynecology and Reproductive Sciences, Integrative Systems Biology Graduate Program, Magee-Womens Research Institute, University of Pittsburgh School of Medicine, Pittsburgh, PA. 3 Cellular and Molecular Biology Program, University of Michigan, Ann Arbor, MI, USA 4 Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI, USA 5 Department of Obstetrics and Gynecology, University of Michigan, Ann Arbor, MI, USA 6Department of Urology, University of Michigan, Ann Arbor, MI, USA 7 These authors contributed equally 8 Lead Contact * Correspondence: [email protected] (K.E.O.), [email protected] (J.Z.L.), [email protected] (S.S.H.) bioRxiv preprint doi: https://doi.org/10.1101/2020.03.17.994509; this version posted March 18, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Summary Spermatogenesis is a highly regulated process that produces sperm to transmit genetic information to the next generation. -

The Centromeric Part of the Human NK Gene Complex: Linkage of LOX-1 and LY49L with the CD94/NKG2 Region
Genes and Immunity (2000) 1, 280–287 2000 Macmillan Publishers Ltd All rights reserved 1466-4879/00 $15.00 www.nature.com/gene The centromeric part of the human NK gene complex: linkage of LOX-1 and LY49L with the CD94/NKG2 region C Bull1, Y Sobanov2,BRo¨hrdanz1, J O’Brien1, H Lehrach1 and E Hofer2 1Max-Planck-Institute for Molecular Genetics, Ihnestrasse 73, D-14195 Berlin, Germany; 2Department of Vascular Biology and Thrombosis Research, Vienna International Research Cooperation Center, University of Vienna, Brunnerstrasse 59, A-1235 Vienna, Austria The natural killer (NK) gene complex is a genomic region containing lectin-type receptor genes. We have established a contig of PAC and BAC clones comprising about 1 Mb of the centromeric part of the NK gene complex. This region extends from the LOX-1 gene, which encodes a receptor for oxidized LDL and was found within 100 kb telomeric of the STS marker D12S77, contains the CD94 and NKG2 NK receptor genes and reaches beyond D12S852 on the proximal side. In this part we have mapped the human LY49L gene, a homologue of the rodent Ly49 genes, which encode important MHC class I receptors for the regulation of NK cell activity in rodents. The LY49L gene is localized 100 to 200 kb centromeric of the NKG2 gene cluster and 300 to 400 kb telomeric of the STS marker D12S841. Genomic sequencing of the complete gene including promoter and intron sequences confirmed that the structure is similar to the mouse Ly49 genes. Screening of several cDNA libraries did not detect any transcripts of putative additional human LY49 genes. -

Gene Section Review
Atlas of Genetics and Cytogenetics in Oncology and Haematology INIST -CNRS OPEN ACCESS JOURNAL Gene Section Review KLRK1 (killer cell lectin-like receptor subfamily K, member 1) Lewis L Lanier UCSF, Department of Microbiology and Immunology, San Francisco, CA 94143-0414, USA (LLL) Published in Atlas Database: June 2014 Online updated version : http://AtlasGeneticsOncology.org/Genes/KLRK1ID41094ch12p13.html DOI: 10.4267/2042/56407 This article is an update of : Lanier LL. KLRK1 (killer cell lectin-like receptor subfamily K, member 1). Atlas Genet Cytogenet Oncol Haematol 2008;12(1):47- 49. This work is licensed under a Creative Commons Attribution-Noncommercial-No Derivative Works 2.0 France Licence. © 2015 Atlas of Genetics and Cytogenetics in Oncology and Haematology Abstract DNA/RNA KLRK1 encodes a type II transmembrane-anchored Note glycoprotein that is expressed as a disulfide-linked KLRK1 is present on chromosome 12 within a homodimer on the surface of Natural Killer (NK) cluster of genes referred to as the "NK complex" cells, gamma/delta TcR+ T cells, CD8+ T cells, and (NKC) because several genes that are preferentially a minor subset of CD4+ T cells. It associates non- expressed by Natural Killer (NK) cells are located covalently with the DAP10 signaling protein and in this region, including on the centromeric side provides activating or costimulatory signals to NK KLRD1 (CD94) and on the telomeric side KLRC4 cells and T cells. NKG2D binds to a family of (NKG2F), KLRC3 (NKG2E), KLRC2 (NKG2C), glycoproteins, in humans the MICA, MICB, and and KLRC1 (NKG2A) (Houchins et al., 1991). ULBP1-6 membrane proteins, which are frequently Description expressed on cells that have become infected with pathogens or undergone transformation. -

Entrez ID Gene Name Fold Change Q-Value Description
Entrez ID gene name fold change q-value description 4283 CXCL9 -7.25 5.28E-05 chemokine (C-X-C motif) ligand 9 3627 CXCL10 -6.88 6.58E-05 chemokine (C-X-C motif) ligand 10 6373 CXCL11 -5.65 3.69E-04 chemokine (C-X-C motif) ligand 11 405753 DUOXA2 -3.97 3.05E-06 dual oxidase maturation factor 2 4843 NOS2 -3.62 5.43E-03 nitric oxide synthase 2, inducible 50506 DUOX2 -3.24 5.01E-06 dual oxidase 2 6355 CCL8 -3.07 3.67E-03 chemokine (C-C motif) ligand 8 10964 IFI44L -3.06 4.43E-04 interferon-induced protein 44-like 115362 GBP5 -2.94 6.83E-04 guanylate binding protein 5 3620 IDO1 -2.91 5.65E-06 indoleamine 2,3-dioxygenase 1 8519 IFITM1 -2.67 5.65E-06 interferon induced transmembrane protein 1 3433 IFIT2 -2.61 2.28E-03 interferon-induced protein with tetratricopeptide repeats 2 54898 ELOVL2 -2.61 4.38E-07 ELOVL fatty acid elongase 2 2892 GRIA3 -2.60 3.06E-05 glutamate receptor, ionotropic, AMPA 3 6376 CX3CL1 -2.57 4.43E-04 chemokine (C-X3-C motif) ligand 1 7098 TLR3 -2.55 5.76E-06 toll-like receptor 3 79689 STEAP4 -2.50 8.35E-05 STEAP family member 4 3434 IFIT1 -2.48 2.64E-03 interferon-induced protein with tetratricopeptide repeats 1 4321 MMP12 -2.45 2.30E-04 matrix metallopeptidase 12 (macrophage elastase) 10826 FAXDC2 -2.42 5.01E-06 fatty acid hydroxylase domain containing 2 8626 TP63 -2.41 2.02E-05 tumor protein p63 64577 ALDH8A1 -2.41 6.05E-06 aldehyde dehydrogenase 8 family, member A1 8740 TNFSF14 -2.40 6.35E-05 tumor necrosis factor (ligand) superfamily, member 14 10417 SPON2 -2.39 2.46E-06 spondin 2, extracellular matrix protein 3437