By Ctpsynthetase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Copyright by Jeremy Daniel O'connell 2012
Copyright by Jeremy Daniel O’Connell 2012 The Dissertation Committee for Jeremy Daniel O’Connell Certifies that this is the approved version of the following dissertation: Systemic Protein Aggregation in Stress and Aging Restructures Cytoplasmic Architecture Committee: Edward Marcotte, Supervisor Dean Appling Andrew Ellington Makkuni Jayaram Scott Stevens Systemic Protein Aggregation in Stress and Aging Restructures Cytoplasmic Architecture by Jeremy Daniel O’Connell, B.S. Dissertation Presented to the Faculty of the Graduate School of The University of Texas at Austin in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy The University of Texas at Austin September 2012 Dedication Cytisus laburnum, simul vincet omnem To my dad and mom who encouraged and enabled my education with countless sacrifices, I promised this graduation would be the one we would attend, and I am truly sorry I was not swift enough to make that possible. Acknowledgements Foremost, I thank my advisor Edward Marcotte, for not just a second lease on a life in science but one in an amazing lab environment. His intellectual rigor, enduring patience, amazing work ethic, and enthusiasm for discovery were an inspiration. I thank my collaborators in this project: Gwen Stovall, Alice Zhao, Gabe Wu, and Mark Tsechansky for their comradery and support on this great adventure. I thank the talented undergraduates: Maguerite West-Driga, Ariel Royall, and Tyler McDonald who stuck with me. Each of you will soon be a better scientist than I ever will, and I hope you enjoyed and learned from our research together nearly as much as I did. -

The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (Cad) by Phosphorylation and Protein-Protein Interactions
THE REGULATION OF CARBAMOYL PHOSPHATE SYNTHETASE-ASPARTATE TRANSCARBAMOYLASE-DIHYDROOROTASE (CAD) BY PHOSPHORYLATION AND PROTEIN-PROTEIN INTERACTIONS Eric M. Wauson A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology. Chapel Hill 2007 Approved by: Lee M. Graves, Ph.D. T. Kendall Harden, Ph.D. Gary L. Johnson, Ph.D. Aziz Sancar M.D., Ph.D. Beverly S. Mitchell, M.D. 2007 Eric M. Wauson ALL RIGHTS RESERVED ii ABSTRACT Eric M. Wauson: The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (CAD) by Phosphorylation and Protein-Protein Interactions (Under the direction of Lee M. Graves, Ph.D.) Pyrimidines have many important roles in cellular physiology, as they are used in the formation of DNA, RNA, phospholipids, and pyrimidine sugars. The first rate- limiting step in the de novo pyrimidine synthesis pathway is catalyzed by the carbamoyl phosphate synthetase II (CPSase II) part of the multienzymatic complex Carbamoyl phosphate synthetase, Aspartate transcarbamoylase, Dihydroorotase (CAD). CAD gene induction is highly correlated to cell proliferation. Additionally, CAD is allosterically inhibited or activated by uridine triphosphate (UTP) or phosphoribosyl pyrophosphate (PRPP), respectively. The phosphorylation of CAD by PKA and ERK has been reported to modulate the response of CAD to allosteric modulators. While there has been much speculation on the identity of CAD phosphorylation sites, no definitive identification of in vivo CAD phosphorylation sites has been performed. Therefore, we sought to determine the specific CAD residues phosphorylated by ERK and PKA in intact cells. -

Classical and Rational Approaches to Antifungal Drug Design
Classical and rational approaches to antifungal drug design Jessica Louise Chitty BSc (Hons) A thesis submitted for the degree of Doctor of Philosophy at The University of Queensland in 2017 School of Chemistry and Molecular Biosciences Institute of Molecular Biosciences Abstract The emergence of human immunodeficiency virus (HIV) in the 1980s has led to an increase in infections from previously rare pathogens. Many of these now cause widespread infection among individuals with compromised immune systems, not just limited to AIDS patients but also to those placed on immunosuppressive medication. The encapsulated yeast Cryptococcus neoformans causes widespread disease in the immunocompromised population, particularly in sub-Saharan Africa where it is a major cause of AIDS-related mortality due in part to limited resources and variable drug availability. Current treatment options are restricted to three out-dated antifungals amphotericin B, flucytosine and fluconazole; where possible they are used in combination as nephrotoxicity and resistance are contributing factors in the unacceptably high mortality rates. Alternative therapeutic agents are urgently required to improve survival rates and combat antifungal drug resistance. Two main routes of compound development can be taken: classical drug screening or rational drug design. Classical design requires groups of compounds to be screened against pathogens and those identified with high efficacy and low cytotoxicity are pursued. Rational drug design requires a detailed characterization of the proposed target; exploitable differences between the pathogen and human host are sought out as potential druggable targets. In this thesis both classical and rational methods have been investigated. A classical approach was taken to investigate a class of octapeptin compounds, produced as secondary metabolites by the soil dwelling bacterium, Bacillus circulans. -

Uva-DARE (Digital Academic Repository)
UvA-DARE (Digital Academic Repository) The pivotal role of CTP synthetase in the metabolism of (deoxy)nucleosides in neuroblastoma Bierau, J. Publication date 2003 Link to publication Citation for published version (APA): Bierau, J. (2003). The pivotal role of CTP synthetase in the metabolism of (deoxy)nucleosides in neuroblastoma. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:27 Sep 2021 1 1 Introduction n "Nucleotidess are water-soluble components which naturally occur, inn larger or smaller portions, in both animal and vegetable foods. Combinedd with other components, theyy are the elements to bring the flavor in food." fromm the Ajinomoto Europe website ChapterChapter 1 Introduction n 1.11 General introduction to neuroblastoma Incidence Incidence Neuroblastomaa is the most common extra cranial solid cancer of childhood. -

1611 REGULATION of PYRIMIDINE METABOLISM in PLANTS Chris
[Frontiers in Bioscience 9, 1611-1625, May 1, 2004] REGULATION OF PYRIMIDINE METABOLISM IN PLANTS 1, 2 1, 3 1, 4 1, 5 1, 6 1, 7 Chris Kafer , Lan Zhou , Djoko Santoso , Adel Guirgis , Brock Weers , Sanggyu Park and Robert Thornburg 1 1 Department of Biochemistry, Biophysics, and Molecular Biology, Iowa State University, Ames, Iowa 50011, 2 BASF Plant Science LLC, 2901 South Loop Drive, Ste 3800, Ames, Iowa 50014, 3 Lan Zhou, Pioneer Hi-Bred International, Inc. 7300 NW 62nd Avenue, PO Box 1004, Johnston, Iowa 50131-1004, 4 Indonesian Biotechnology Research Institute for Estate Crops, Jl, Taman Kencana No 1, Bogor 16151 Indonesia, 5 Institute of Genetic Engineering and Biotechnology, Menofiya University, PO Box 79/22857, Sadat City, Egypt, 6 Department of Biochemistry, University of Iowa, 4/511 Bowen Science Building, Iowa City, Iowa 52242-1109, 7 Division of Life and Environment, College of Natural Resources, Daegu University, Gyongsan City, Gyongbuk, Korea 712-714 TABLE OF CONTENTS 1. Abstract 2. Introduction 3. Pyrimidine metabolic pathways 3.1. De novo pyrimidine biosynthesis 3.1.1. CPSase 3.1.2. ATCase 3.1.3. DHOase 3.1.4. DHODH 3.1.5. UMPS 3.1.6. Intracellular Organization of the de novo Pathway 3.2. Pyrimidine Salvage and Recycling 3.2.1. Cytosine deaminase 3.2.2. Cytidine deaminase 3.2.3. UPRTase 3.3. Pyrimidine Modification 3.3.1. UMP/CMP kinase 3.3.2. NDP kinase 3.3.3. CTP synthase, NDP reductase, dUTPase 3.3.4. Thymidylate synthase/Dihydrofolate reductase 3.4. Pyrimidine Catabolism 4. Regulation of pyrimidine metabolism 4.1. -

Developmental Disorder Associated with Increased Cellular Nucleotidase Activity (Purine-Pyrimidine Metabolism͞uridine͞brain Diseases)
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 11601–11606, October 1997 Medical Sciences Developmental disorder associated with increased cellular nucleotidase activity (purine-pyrimidine metabolismyuridineybrain diseases) THEODORE PAGE*†,ALICE YU‡,JOHN FONTANESI‡, AND WILLIAM L. NYHAN‡ Departments of *Neurosciences and ‡Pediatrics, University of California at San Diego, La Jolla, CA 92093 Communicated by J. Edwin Seegmiller, University of California at San Diego, La Jolla, CA, August 7, 1997 (received for review June 26, 1997) ABSTRACT Four unrelated patients are described with a represent defects of purine metabolism, although no specific syndrome that included developmental delay, seizures, ataxia, enzyme abnormality has been identified in these cases (6). In recurrent infections, severe language deficit, and an unusual none of these disorders has it been possible to delineate the behavioral phenotype characterized by hyperactivity, short mechanism through which the enzyme deficiency produces the attention span, and poor social interaction. These manifesta- neurological or behavioral abnormalities. Therapeutic strate- tions appeared within the first few years of life. Each patient gies designed to treat the behavioral and neurological abnor- displayed abnormalities on EEG. No unusual metabolites were malities of these disorders by replacing the supposed deficient found in plasma or urine, and metabolic testing was normal metabolites have not been successful in any case. except for persistent hypouricosuria. Investigation of purine This report describes four unrelated patients in whom and pyrimidine metabolism in cultured fibroblasts derived developmental delay, seizures, ataxia, recurrent infections, from these patients showed normal incorporation of purine speech deficit, and an unusual behavioral phenotype were bases into nucleotides but decreased incorporation of uridine. -

Supplementary Informations SI2. Supplementary Table 1
Supplementary Informations SI2. Supplementary Table 1. M9, soil, and rhizosphere media composition. LB in Compound Name Exchange Reaction LB in soil LBin M9 rhizosphere H2O EX_cpd00001_e0 -15 -15 -10 O2 EX_cpd00007_e0 -15 -15 -10 Phosphate EX_cpd00009_e0 -15 -15 -10 CO2 EX_cpd00011_e0 -15 -15 0 Ammonia EX_cpd00013_e0 -7.5 -7.5 -10 L-glutamate EX_cpd00023_e0 0 -0.0283302 0 D-glucose EX_cpd00027_e0 -0.61972444 -0.04098397 0 Mn2 EX_cpd00030_e0 -15 -15 -10 Glycine EX_cpd00033_e0 -0.0068175 -0.00693094 0 Zn2 EX_cpd00034_e0 -15 -15 -10 L-alanine EX_cpd00035_e0 -0.02780553 -0.00823049 0 Succinate EX_cpd00036_e0 -0.0056245 -0.12240603 0 L-lysine EX_cpd00039_e0 0 -10 0 L-aspartate EX_cpd00041_e0 0 -0.03205557 0 Sulfate EX_cpd00048_e0 -15 -15 -10 L-arginine EX_cpd00051_e0 -0.0068175 -0.00948672 0 L-serine EX_cpd00054_e0 0 -0.01004986 0 Cu2+ EX_cpd00058_e0 -15 -15 -10 Ca2+ EX_cpd00063_e0 -15 -100 -10 L-ornithine EX_cpd00064_e0 -0.0068175 -0.00831712 0 H+ EX_cpd00067_e0 -15 -15 -10 L-tyrosine EX_cpd00069_e0 -0.0068175 -0.00233919 0 Sucrose EX_cpd00076_e0 0 -0.02049199 0 L-cysteine EX_cpd00084_e0 -0.0068175 0 0 Cl- EX_cpd00099_e0 -15 -15 -10 Glycerol EX_cpd00100_e0 0 0 -10 Biotin EX_cpd00104_e0 -15 -15 0 D-ribose EX_cpd00105_e0 -0.01862144 0 0 L-leucine EX_cpd00107_e0 -0.03596182 -0.00303228 0 D-galactose EX_cpd00108_e0 -0.25290619 -0.18317325 0 L-histidine EX_cpd00119_e0 -0.0068175 -0.00506825 0 L-proline EX_cpd00129_e0 -0.01102953 0 0 L-malate EX_cpd00130_e0 -0.03649016 -0.79413596 0 D-mannose EX_cpd00138_e0 -0.2540567 -0.05436649 0 Co2 EX_cpd00149_e0 -

University of Cincinnati
UNIVERSITY OF CINCINNATI Date: 22-Jan-2010 I, Amruta Desai , hereby submit this original work as part of the requirements for the degree of: Master of Science in Computer Engineering It is entitled: Design support for biomolecular systems Student Signature: Amruta Desai This work and its defense approved by: Committee Chair: Carla Purdy, C, PhD Carla Purdy, C, PhD Wen-Ben Jone, PhD Wen-Ben Jone, PhD George Purdy, PhD George Purdy, PhD 2/2/2010 389 Design Support for Biomolecular Systems A thesis submitted to the Division of Graduate Research and Advanced Studies of The University of Cincinnati In partial fulfillment of the Requirements for the degree of Master of Science in the Department of Electrical and Computer Engineering of the College of Engineering By Amruta Desai BE in Electrical Engineering, Rajiv Gandhi Technical University, 2005 January, 2010 Thesis Advisor and Committee Chair: Dr. Carla Purdy Abstract Systems biology is an emerging field which connects system level understanding to molecular level understanding. Biomolecular systems provide a comprehensive view of a biological phenomenon, in the form of a network of inter-related reactions or processes. The work described in this thesis focuses on developing the support for virtual experiments in systems biology. This will help biologists to make choices about which wet lab experiments are likely to be the most informative, thereby saving both time and material resources. Our goal is to support synthetic biology by providing tools which can be employed by biologists, engineers, and computational scientists. Our approach makes use of well-developed techniques from the field of VLSI design. -

Active Site Coupling in Plasmodium Falciparum GMP Synthetase Is Triggered by Domain Rotation
ARTICLE Received 7 May 2015 | Accepted 19 Oct 2015 | Published 23 Nov 2015 DOI: 10.1038/ncomms9930 OPEN Active site coupling in Plasmodium falciparum GMP synthetase is triggered by domain rotation Lionel Ballut1,*, Se´bastien Violot1,*, Santosh Shivakumaraswamy2,*, Lakshmi Prasoona Thota2, Manu Sathya2, Jyothirmai Kunala2, Bauke W. Dijkstra3, Raphae¨l Terreux4, Richard Haser1, Hemalatha Balaram2 & Nushin Aghajari1 GMP synthetase (GMPS), a key enzyme in the purine biosynthetic pathway performs catalysis through a coordinated process across two catalytic pockets for which the mechanism remains unclear. Crystal structures of Plasmodium falciparum GMPS in conjunction with mutational and enzyme kinetic studies reported here provide evidence that an 85° rotation of the GATase domain is required for ammonia channelling and thus for the catalytic activity of this two-domain enzyme. We suggest that conformational changes in helix 371–375 holding catalytic residues and in loop 376–401 along the rotation trajectory trigger the different steps of catalysis, and establish the central role of Glu374 in allostery and inter- domain crosstalk. These studies reveal the mechanism of domain rotation and inter-domain communication, providing a molecular framework for the function of all single polypeptide GMPSs and form a solid basis for rational drug design targeting this therapeutically important enzyme. 1 BioCrystallography and Structural Biology of Therapeutic Targets Group, Molecular and Structural Bases of Infectious Systems, UMR5086 CNRS-University of Lyon 1, 7 passage du Vercors, 69367 Lyon Cedex 07, France. 2 Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Jakkur, Bangalore 560064, India. 3 Laboratory of Biophysical Chemistry, University of Groningen, Nijenborgh 7, 9747 AG Groningen, The Netherlands. -

Staphylococcus Aureus Infection Reduces Nutrition Uptake and Nucleotide Biosynthesis in a Human Airway Epithelial Cell Line
H OH metabolites OH Article Staphylococcus aureus Infection Reduces Nutrition Uptake and Nucleotide Biosynthesis in a Human Airway Epithelial Cell Line Philipp Gierok 1, Manuela Harms 2, Karen Methling 1, Falko Hochgräfe 2 and Michael Lalk 1,* 1 Institute of Biochemistry, University of Greifswald, 17487 Greifswald, Germany; [email protected] (P.G.); [email protected] (K.M.) 2 Competence Center Functional Genomics, Junior Research Group Pathoproteomics, University of Greifswald, 17487 Greifswald, Germany; [email protected] (M.H.); [email protected] (F.H.) * Correspondence: [email protected]; Tel.: +49-3834-86-4867 Academic Editors: Wolfgang Eisenreich and Adelbert Bacher Received: 6 October 2016; Accepted: 2 November 2016; Published: 9 November 2016 Abstract: The Gram positive opportunistic human pathogen Staphylococcus aureus induces a variety of diseases including pneumonia. S. aureus is the second most isolated pathogen in cystic fibrosis patients and accounts for a large proportion of nosocomial pneumonia. Inside the lung, the human airway epithelium is the first line in defence with regard to microbial recognition and clearance as well as regulation of the immune response. The metabolic host response is, however, yet unknown. To address the question of whether the infection alters the metabolome and metabolic activity of airway epithelial cells, we used a metabolomics approach. The nutrition uptake by the human airway epithelial cell line A549 was monitored over time by proton magnetic resonance spectroscopy (1H-NMR) and the intracellular metabolic fingerprints were investigated by gas chromatography and high performance liquid chromatography (GC-MS) and (HPLC-MS). To test the metabolic activity of the host cells, glutamine analogues and labelled precursors were applied after the infection. -

Metabolite Name Chemical Formula KEGG/HMDB/ Pubchem Identifier Glyoxylate C2H2O3 C00048 Glycolate C2H4O3 C00160 Pyruvate C3H4O3
Metabolite name Chemical formula KEGG/HMDB/ PubChem Identifier glyoxylate C2H2O3 C00048 glycolate C2H4O3 C00160 pyruvate C3H4O3 C00022 lactate C3H6O3 C00186 2-oxobutanoate C4H6O3 C00109 acetoacetate C4H6O3 C00164 glycerate C3H6O4 C00258 uracil C4H4N2O2 C00106 fumarate C4H4O4 C00122 Maleic acid C4H4O4 C01384 2-keto-isovalerate C5H8O3 C00141 Guanidoacetic acid C3H7N3O2 C00581 succinate C4H6O4 C00042 Methylmalonic acid C4H6O4 C02170 3-S-methylthiopropionate C4H8O2S C08276 nicotinate C6H5NO2 C00253 taurine C2H7NO3S C00245 Pyroglutamic acid C5H7NO3 C01879 Citraconic acid C5H6O4 C02226 2-ketohaxanoic acid C6H10O3 HMDB01864 N-Acetyl-L-alanine C5H9NO3 C01073 oxaloacetate C4H4O5 C00036 Hydroxyisocaproic acid C6H12O3 HMDB00746 malate C4H6O5 C00149 hypoxanthine C5H4N4O C00262 anthranilate C7H7NO2 C00108 p-aminobenzoate C7H7NO2 C00568 p-hydroxybenzoate C7H6O3 C00156 acetylphosphate C2H5O5P C00227 Carbamoyl phosphate CH4NO5P C00169 a-ketoglutarate C5H6O5 C00026 Phenylpropiolic acid C9H6O2 HMDB00563 2-oxo-4-methylthiobutanoate C5H8O3S C01180 2-Hydroxy-2-methylbutanedioic acid C5H8O5 C02612 3-methylphenylacetic acid C9H10O2 HMDB02222 xanthine C5H4N4O2 C00385 Hydroxyphenylacetic acid C8H8O3 C05852 2,3-dihydroxybenzoic acid C7H6O4 C00196 orotate C5H4N2O4 C00295 dihydroorotate C5H6N2O4 C00337 allantoin C4H6N4O3 C01551 Aminoadipic acid C6H11NO4 C00956 Indole-3-carboxylic acid C9H7NO2 HMDB03320 phenylpyruvate C9H8O3 C00166 Atrolactic acid C9H10O3 HMDB00475 Phenyllactic acid C9H10O3 C01479 quinolinate C7H5NO4 C03722 phosphoenolpyruvate C3H5O6P C00074 Uric -
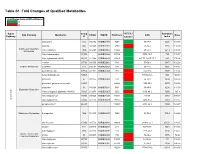
Table S1. Fold Changes of Qualified Metabolites
Table S1. Fold Changes of Qualified Metabolites Fold Change: Cases of WTC-LI/Control <1 >1 Super Comp WTC-LI Retention Sub Pathway Metabolite KEGG HMDB PubChem CAS Mass Pathway ID Control Index asparagine 512 C00152 HMDB00168 6267 0.99 70-47-3 1225 133.06 aspartate 443 C00049 HMDB00191 5960 1.09 56-84-8 1165 134.04 Alanine and Aspartate N-acetylalanine 1585 C02847 HMDB00766 88064 0.99 97-69-8 861.2 130.05 Metabolism N-acetylasparagine 33942 HMDB06028 99715 0.82 4033-40-3 785 175.07 N-acetylaspartate (NAA) 22185 C01042 HMDB00812 65065 0.95 997-55-7;997-55-7 880 176.06 creatine 27718 C00300 HMDB00064 586 0.95 57-00-1 1947 132.08 Creatine Metabolism creatinine 513 C00791 HMDB00562 588 0.99 60-27-5 2055 114.07 guanidinoacetate 43802 C00581 HMDB00128 763 1.05 352-97-6 1937 118.06 beta-citrylglutamate 54923 1.05 73590-26-8 900 322.08 glutamate 57 C00025 HMDB00148 611 1.06 56-86-0 1500 148.06 glutamate, gamma-methyl ester 33487 68662 0.89 1499-55-4 2170 162.08 glutamine 53 C00064 HMDB00641 5961 0.89 56-85-9 1291 147.08 Glutamate Metabolism N-acetyl-aspartyl-glutamate (NAAG) 35665 C12270 HMDB01067 5255 1.04 3106-85-2 1035 305.1 N-acetylglutamate 15720 C00624 HMDB01138 70914 0.84 8/3/17 1050 190.07 Amino Acid Amino N-acetylglutamine 33943 C02716 HMDB06029 182230 0.48 2490-97-3 2140 187.07 pyroglutamine* 46225 134508 0.89 2353-44-8 1900 129.07 Glutathione Metabolism 5-oxoproline 1494 C01879 HMDB00267 7405 0.93 98-79-3 738.5 128.04 allo-threonine 15142 C05519 HMDB04041 99289 0.91 28954-12-3 2511.1 118.05 betaine 3141 C00719 HMDB00043 247 0.92 107-43-7