Crop Science
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

C a Se Stud Y
This project is funded by the European Union November 2020 Culture in ruins The illegal trade in cultural property Case study: Algeria and Tunisia Julia Stanyard and Rim Dhaouadi Summary This case study forms part of a set of publications on the illegal trade in cultural property across North and West Africa, made up of a research paper and three case studies (on Mali, Nigeria and North Africa). This study is focused on Algeria and Tunisia, which share the same forms of material culture but very different antiquity markets. Attention is given to the development of online markets which have been identified as a key threat to this region’s heritage. Key findings • The large-scale extraction of cultural objects in both countries has its roots in the period of French colonial rule. • During the civil war in Algeria in the 1990s, trafficking in cultural heritage was allegedly linked to insurgent anti-government groups among others. • In Tunisia, the presidential family and the political elite reportedly dominated the country’s trade in archaeological objects and controlled the illegal markets. • The modern-day trade in North African cultural property is an interlinked regional criminal economy in which objects are smuggled between Tunisia and Algeria as well as internationally. • State officials and representatives of cultural institutions are implicated in the Algerian and Tunisian antiquities markets in a range of different capacities, both as passive facilitators and active participants. • There is evidence that some architects and real estate entrepreneurs are connected to CASE STUDY CASE trafficking networks. Introduction The region is a palimpsest of ancient material,7 much of which remains unexplored and unexcavated by Cultural heritage in North Africa has come under fire archaeologists. -

Dzgrid Initiative GRID Nationale
Algerian Research Network ARN Aouaouche El-Maouhab Manager of Algerian Research Network ARN [email protected] ARN - Connectivity National backbone based on 10 PoPs International Connectivity through : GEANT (European Research Network) with 2.5 Gbps , upgraded since Junuary 2016 under EC AfricaConnect2 cluster 3 project Internet commodity with 1 Gbps Mbps shortly upgraded to 2 Gbps « ARN & DZ e-Science GRID » (GEANT2) ARN Map STM16 Internet MESRS,DGRSDT,UMBB, UNIV. ALGER (Ben-Aknoun1, Ben-Aknoun2, Bouzareah, Beni-Messous, Dely-Brahim, Maherzi, Kharrouba,Dergana), ENTP, STM4+STM1 USTHB, ENP,ESI, ENV,ENTP,ENSH,INA,EPAU,ISMAL,INPS,INC,ESC,ENS(KOUBA), ENS(BOUZAREAH),CERIST,CDTA,CDER,CSC,CRSTDLA,CREAD,CRAPC,UDTS,UDES ANDRU,ANVREDET,UFC,INRAA,CGS,ENA,CRAAG,INFS/STS,CRNB,INRE INRAA, CGS, ENA, CRAAG, U. Constantine, U. Emir AEK, ENS, CRBiotech., INFS/STS, CRNB, INRE Bejaia U. ES-SENIA,USTO,ENST,CRASC ANDRS, El-Tarf Mila Guelma Mostaganem Bordj Bou Arreridj Oum-El-Bouaghi Tissemsilt Sidi Bel Abbes (U. Biskra,CRSTRA) (U. Ghardaia, URAER) Adrar Nouveau PoP (U. Adrar, UEES) Lien GE STM4 Tamanrasset « STM1La grille nationale DZ eScience GRID » FE 100M Oran, 30 Mai 2012 FE 10M E-Infrastructure in Algeria e-Science Collaborations DZ e-Science VO National GRID VOs DZ e-Science GRID Distributed Computing National GRID Infrastructure Infrastructure ARN Network Infrastructure Academic & Research Network « ARN & DZ e-Science GRID » DZ e-Science GRID infrastructure Core services Task manager WMS Monitoring + VO manager + DZ e-Science CA VMProxy user supp. -

Potential Threats to Afro-Palearctic Migrato
www.nature.com/scientificreports OPEN Unravelling the drastic range retraction of an emblematic songbird of North Africa: potential Received: 31 October 2016 Accepted: 16 March 2017 threats to Afro-Palearctic migratory Published: xx xx xxxx birds Rassim Khelifa1, Rabah Zebsa2, Hichem Amari3, Mohammed Khalil Mellal4, Soufyane Bensouilah3, Abdeldjalil Laouar5 & Hayat Mahdjoub1 Understanding how culture may influence biodiversity is fundamental to ensure effective conservation, especially when the practice is local but the implications are global. Despite that, little effort has been devoted to documenting cases of culturally-related biodiversity loss. Here, we investigate the cultural domestication of the European goldfinch (Carduelis carduelis) in western Maghreb (Morocco, Algeria and Tunisia) and the effects of long-term poaching of wild populations (1990–2016) on range distribution, socio-economic value, international trading and potential collateral damage on Afro- Palearctic migratory birds. On average, we found that the European goldfinch lost 56.7% of its distribution range in the region which led to the increase of its economic value and establishment of international trading network in western Maghreb. One goldfinch is currently worth nearly a third of the average monthly income in the region. There has been a major change in poaching method around 2010, where poachers started to use mist nets to capture the species. Nearly a third of the 16 bird species captured as by-catch of the European goldfinch poaching are migratory, of which one became regularly sold as cage-bird. These results suggest that Afro-Palearctic migratory birds could be under serious by-catch threat. Species overexploitation for wildlife trade is a major global threat to biodiversity, particularly birds1, 2. -

Download Download
Nisan / The Levantine Review Volume 4 Number 2 (Winter 2015) Identity and Peoples in History Speculating on Ancient Mediterranean Mysteries Mordechai Nisan* We are familiar with a philo-Semitic disposition characterizing a number of communities, including Phoenicians/Lebanese, Kabyles/Berbers, and Ismailis/Druze, raising the question of a historical foundation binding them all together. The ethnic threads began in the Galilee and Mount Lebanon and later conceivably wound themselves back there in the persona of Al-Muwahiddun [Unitarian] Druze. While DNA testing is a fascinating methodology to verify the similarity or identity of a shared gene pool among ostensibly disparate peoples, we will primarily pursue our inquiry using conventional historical materials, without however—at the end—avoiding the clues offered by modern science. Our thesis seeks to substantiate an intuition, a reading of the contours of tales emanating from the eastern Mediterranean basin, the Levantine area, to Africa and Egypt, and returning to Israel and Lebanon. The story unfolds with ancient biblical tribes of Israel in the north of their country mixing with, or becoming Lebanese Phoenicians, travelling to North Africa—Tunisia, Algeria, and Libya in particular— assimilating among Kabyle Berbers, later fusing with Shi’a Ismailis in the Maghreb, who would then migrate to Egypt, and during the Fatimid period evolve as the Druze. The latter would later flee Egypt and return to Lebanon—the place where their (biological) ancestors had once dwelt. The original core group was composed of Hebrews/Jews, toward whom various communities evince affinity and identity today with the Jewish people and the state of Israel. -

Chemical Composition and Antioxidant, Anti-Inflammatory, And
molecules Article Chemical Composition and Antioxidant, Anti-Inflammatory, and Enzyme Inhibitory Activities of an Endemic Species from Southern Algeria: Warionia saharae Habiba Rechek 1,2,3 , Ammar Haouat 4,5, Kaouther Hamaidia 1,6,* , Hamza Allal 7 , Tarek Boudiar 8, Diana C. G. A. Pinto 3,* , Susana M. Cardoso 3 , Chawki Bensouici 8, Noureddine Soltani 6 and Artur M. S. Silva 3,* 1 Faculty of Sciences of Nature and Life, Mohamed Cherif Messaadia University, Souk-Ahras 41000, Algeria; [email protected] 2 Department of Biology of Organisms, Faculty of Sciences of Nature and Life, University of Batna 2, Mostefa Ben Boulaid, Batna 05078, Algeria 3 LAQV-REQUIMTE & Department of Chemistry, University of Aveiro, 3810-193 Aveiro, Portugal; [email protected] 4 Unité de Valorisation des Ressources Naturelles, Molécules Bioactives et Analyse Physicochimiques et Biologiques (VARENBIOMOL), Université des Frères Mentouri, Constantine 25000, Algeria; [email protected] 5 Department of Biology, Faculty of Sciences of Nature and Life, University of Oued Souf, Oued Souf 39000, Algeria 6 Laboratory of Applied Animal Biology, Badji Mokhtar University, Annaba 23000, Algeria; [email protected] Citation: Rechek, H.; Haouat, A.; 7 Department of Technology, Faculty of Technology, 20 August 1955 Skikda University, Hamaidia, K.; Allal, H.; Boudiar, T.; Skikda 21000, Algeria; [email protected] Pinto, D.C.G.A.; Cardoso, S.M.; 8 Centre de Recherche en Biotechnologie, Ali Mendjli Nouvelle Ville UV 03, Constantine 25000, Algeria; Bensouici, C.; Soltani, N.; Silva, [email protected] (T.B.); [email protected] (C.B.) A.M.S. Chemical Composition and * Correspondence: [email protected] (K.H.); [email protected] (D.C.G.A.P.); Antioxidant, Anti-Inflammatory, and [email protected] (A.M.S.S.); Tel.: +213-66-509-5858 (K.H.); +351-234-401407 (D.C.G.A.P.); Enzyme Inhibitory Activities of an +351-234-370714 (A.M.S.S.) Endemic Species from Southern Algeria: Warionia saharae. -

Liste Des Societe D'expertise Et Experts Agrees Par L'uar "Jijel"
01, Lot Said HAMDINE, Bir Mourad Rais, - Alger - BP 226 CP 16033, ALGER. Tél. : (213) (0) 21 54 74 96 & 98 Fax : (213) (0) 21 54 69 22 Site Web : www.uar.dz - e-mail : [email protected] Association régie par l’ordonnance 95/07 du 25/01/1995 modifiée et complétée. LISTE DES SOCIETE D'EXPERTISE ET EXPERTS AGREES PAR L'UAR "JIJEL" Date N° Nom et Prénom Adresse Professionnelle Spécialité Tel. Mobile Fax E-Mail d'inscription EURL COSEANAV Cité 112 Logts EPLF Plage, Bt 07, (034) 47 36 67 (0560) 061 166 [email protected] 1 Local B, Jijel Facultés maritimes 04/06/2017 BOUTAOUI Omar Route Nationale N° 43, Rue Bâtiment 26/02/2013 (034) 47 02 91 (0661) 636 596 (034) 47 02 91 [email protected] 2 Mohamed Boutaghou, N° 16 A, El Aouana Centre, Jijel 3 BOUKEDIRA Messaoud Cité Zaâmouche Taher, Jijel Bâtiment 27/07/2005 (034) 44 02 30 (0775) 552 637 BENZAID Thaki-Eddine Lot khellaf, Rue El Moudjahiddine, Bâtiment 26/02/2013 (034) 47 44 64 (0770) 249 800 4 Local N° A, Jijel 5 ALLAOUA Hamanou 7, Rue Larbi Ben M`hidi, Jijel Agronomie 03/05/2004 (034) 49 41 01 ALLEL Ahmed Tassoust Commune Emir Agronomie 01/01/1900 (034) 51 02 52 (0661) 335 896 6 Abdelkader, Jijel 7 TIBOUK Saida Rue Boltamine Ferhat, Taher, Jijel Agronomie 26/09/2011 (0774) 946 160 AMAROUAYACHE Abdelbaki Cité Aissa Harièche, Crête Est, 150 Agronomie 02/06/2002 (034) 47 65 82 8 logts, Bt C2, Appt N° 9, Jijel ZITOUNI Essaadi Lotissement Zone de stockage, N° Automobile 24/10/2012 (0773) 321 359 [email protected] 9 47, Local N° 48 B, Jijel BELAL Ayache 86, Avenue Abdelhamid Ben-Badis, -

Curriculum Vitae
Curriculum vitae Name: Lazhari BOUZID DOB: 01/01/1953 Marital status: Married Phone: Home: 00 213 31 90 90 45 Cell: 00213 661 53 45 92 Office: 00 213 21 73 61 14 Fax: 00 213 21 73 62 91 Email: [email protected] Address: Home: Rue Sahraoui Belgacem, N 731, El - Fedj, Ain El Bey 2, Constantine – 25020, Algeria Office: Conseil de la Nation, 7 Boulevard Zighoud Youcef, Algiers – 16000, Algeria Current position: - Senator (Algerian Council of the Nation) - Member of the Foreign Affairs Commission, at the Algerian Council of the Nation Working languages: - Arabic - English - French Educational background: - 1976: Bachelor of Law, University of Constantine, Algeria. - 1978: Diploma in Public International Law, University of London, UK - 1979: LL.M in public international law, university of London, UK - 1990: PhD in International Law, University of Glasgow, UK Main Professional activities: - June 2007 to present: Member of the Foreign Affairs Commission, Council of the Nation - April 2007: Nominated for a third term as Senator—the term will end in January 2013 - October 2006 to present: President of the Algerian-British Parliamentary Friendship Group - December 2005: President of the National Liberation Front party’s (FLN) Commission in charge of revising the constitution and the laws on elections and political parties - 2004-2007: Member of the Defense Commission, Council of the Nation - February 2004: Nomination for a second term as Senator - 2003-2004: Member of the Commission in charge of drafting the Presidential programme for the 2004 -
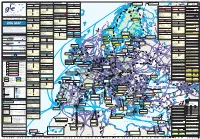
Storage Map Lng
80.25 4.90 Petrozavodsk 39.25 7079 372 082 Huntorf L-gas N 3020 AUSTRIA 65.70 3.50 31.20 LATVIA 2320 SLOVAKIA 31.35 1071 Belgium, Zeebrugge 19.80 3.30Spain, Barcelona Turkey,11.64 Aliaga UK, Milford Haven SHTOKMAN UGS Láb complex 32.40 001 Schönkirchen / Reyersdorf 1544 236 N 083 Nüttermoor L-gas N Norway, Snøhvit Russia, Shtokman Pechora129 SeaIncukalns 2320 R 155 Russia, Yamal2365 LNG N Start-up : 1987 13.50 2.10Start-up : 1968 Start-up : 2006 South Hook LNG SNØHVIT (incl. Gajary baden) 24.50 ASKELADD Melkøya / Hammersfest Teriberka - Barents Sea Yamal Peninsula, north of Western Siberia 3.30 0.50 17.52 6.85 002 Tallesbrunn 347 53 N 084 Nüttermoor H-gas 541 N Start-up : 2009 MELKØYA Incukalns 3200 156 655 N 2.60 0.40 9.96 ALBATROSS Start-up : August 2007 Start-up : 2019 Láb IV 6.85 MAX. HOURLY CAPACITY NOM. ANNUAL CAPACITY MAX. HOURLY CAPACITY NOM. ANNUAL CAPACITY MAX. HOURLY CAPACITY NOM. ANNUAL CAPACITY Hammerfest Petrozavodsk Start-up : 2017 3 2.703 0.40 3 Nüttermoor-H : Storage3 Capacity connected to the Netherlands3 (GTS) 3 Salekhard 003 currentThann : 1.700.000 m (N)/h current217 : 9 bm (N)/year33 1.950.000N m (N)/h 17,1 bmE.ON (N)/year share of 109,3mcm solely used680.000 in NL m (N)/h 6,0 bm (N)/year MAX. HOURLY CAPACITY NOM. ANNUAL CAPACITY 31.50 2.40 0.30 3 3 LITHUANIA SPAIN 4103 3 3 3.36 2.440.000 m (N)/h 21 bm (N)/year 22.70 by 2015 : 2.150.000 m (N)/h by 2015 : 12 bm (N)/year2.88 085 Rüdersdorf H-gas 135 N 004 16. -

The Epigraphy of the Tophet
ISSN 2239-5393 The Epigraphy of the Tophet Maria Giulia Amadasi Guzzo – José Ángel Zamora López (Sapienza Università di Roma – CSIC, Madrid) Abstract The present contribution reassesses the main aspects of the epigraphic sources found in the so-called tophet in order to demonstrate how they are significant and how they undermine the funerary interpretations of these precincts. The inscriptions decisively define the tophet as a place of worship, a sanctuary where sacrifices were made to specific deities in specific rites. The epigraphic evidence combined with literary and archaeological data show how these sacrifices consisted of infants and small animals (either as substitutes or interred together), sometimes commemorated by the inscriptions themselves. Keywords History of Religions, Child Sacrifice, Northwest Semitic Epigraphy, Mediterranean History, Phoenician & Punic World. 1. Introduction Our basic knowledge of the special type of Phoenician and Punic sanctuaries called tophet (a conventional term taken from the Hebrew Bible) seems to be based on wide variety of sources that can be combined to provide an overall interpretation. In fact, archaeological research now provides us with relatively substantial knowledge of the geographical and chronological distribution of these sacred sites and of their structure. Present in some central Mediterranean Phoenician settlements (including on Sardinia) from their foundation, or shortly after, they persist and multiply in North Africa at a later period, generally after the destruction of Carthage1. Archaeology, also, enables us to formulate a “material” definition of these places: they are always– essentially – open-air sites constantly located on the margins of towns, where pottery containers are buried in which the burnt remains of babies and/or baby Received: 11.09.2013. -

Communes Vertes » Gestion Énergétique Durable Des Communes
Ph : © GIZ Mis en oeuvre par: Ministère de l’Intérieur, des Collectivités Locales et de l'Aménagement du Territoire Projet « Communes Vertes » Gestion énergétique durable des communes Contexte Nom du projet Communes Vertes De par ses engagements internationaux dans le cadre des né- gociations sur le climat, et répondant aux défis climatiques lo- Sur mandat du Ministère fédéral de la Coopération Econo- mique et du Développement (Bundesminis- caux, l’Algérie s’est engagée dans un programme ambitieux terium für wirtschaftliche Zusammenarbeit pour contribuer à la réduction des émissions de gaz à effet de und Entwicklung - BMZ) serre (GES) visant 7 à 20% à l’horizon 2030 en comparaison Partenaire politique Le Ministère de l'Intérieur, des Collectivités avec un scénario de consommation classique. locales et de l'Aménagement du territoire (MICLAT) Le développement des énergies renouvelables (EnR) et la pro- motion de l’efficacité énergétique (EE) est un axe principal de Organe d’exécution Deutsche Gesellschaft für Internationale Zu- la stratégie du gouvernement algérien pour atteindre ses ob- sammenarbeit (GIZ) GmbH jectifs de réduction des émissions GES. Cela passe à travers Zones d’intervention Adrar, Bechar, Djelfa, Jijel, Mascara, Msila, l’optimisation de la consommation d'énergie, et en assurant Relizane, Souk Ahras une transition énergétique basée sur la production d’énergie Durée 04/2020 – 03/2023 propre et durable à partir de sources d'EnR. Une attention particulière est mise sur l'électricité provenant de l'énergie Dans ce contexte, le présent projet financé par le Ministère solaire. fédéral Allemand de la Coopération Économique et du Déve- Les collectivités locales jouent un rôle important dans la vi- loppement (BMZ) vient pour soutenir les efforts engagés par sion du gouvernement algérien pour assurer la transition le MICLAT pour la promotion de l’utilisation des EnR et de l’EE énergétique. -

RAPPORT DE SITUATION SUR L'epidemie DU COVID-19 En Algérie Contexte
RAPPORT DE SITUATION SUR L’EPIDEMIE DU COVID-19 en Algérie Date de début Le premier cas positif a été déclaré le 25 février 2020 Rapport N° 110 Date du rapport : 10 Juillet 2020 Date des Données 09 Juillet 2020 à 16H Quatre cent soixante (460) nouveaux cas de COVID-19 ont été notifiés le 09 juillet 2020 portant le total des cas à 17 808 depuis le début de l’épidémie ; Dix (10) nouveaux décès ont été notifiés ce jour portant le total à neuf cent quatre-vingt- huit (988) décès de cas confirmés depuis le début de l’épidémie (létalité des cas confirmés par PCR : 5,55%) ; Dix (10) wilayas sur les 48 n’ont pas notifié de nouveaux cas confirmés pendant les dernières 24 heures ; Trois cent huit (308) patients parmi les cas confirmés ont été sortis de l’hôpital guérit ce jour après des tests de contrôle négatifs portant le nombre total des patients sortis de l’hôpital depuis le début de l’épidémie à 12 637 ; Cinquante-trois (53) patients COVID-19 sont sous assistance respiratoire dans les services de soins intensifs sur l’ensemble du pays ; Maintien du confinement jusqu’au 13 juillet 2020 pour 29 wilayas dont Alger et Blida avec aménagement des horaires de 20h00 à 05h00 du matin. Interdiction pour une semaine, à compter du 10 juillet 2020, de la circulation routière, y compris des véhicules particuliers, de et vers les 29 wilayas suivantes: Boumerdes, Souk Ahras, Tissemsilt, Djelfa, Mascara, Oum El Bouaghi, Batna, Bouira, Relizane, Biskra, Khenchela, M’sila, Chlef, Sidi Bel Abbes, Médéa, Blida, Bordj Bou Arreridj, Tipaza, Ouargla, Bechar, Alger, Constantine, Oran, Sétif, Annaba, Bejaia, Adrar, Laghouat et El Oued ; Interdiction, à compter du vendredi 10 juillet 2020, du transport urbain public et privé durant les week-ends au niveau des 29 wilayas impactées. -

Physical Preparation and Performance Analysis Technology
The Conference Organizing Committee : The Scientific Committee of the Conference: The Chairman of the Conference Organizing People’s Democratic and Republic of Algeria The Chairman of the Scientific Committee of the Committee: Dr. GHERIBI Hichem Ministry of High Education and Scientific Research Conference: Pr. GHENNAM Noureddine University of L'arbi Ben M'hidi Oum El Bouaghi Pr. IDIR Hassan University of Oum El Bouaghi Institute for Sciences and Techniques of Pr.GUELLATI Yazid University of Oum El Bouaghi Dr.CHELIHI Omar : Physical and Sports Activities Pr. Nouasria Mouna University of Oum El Bouaghi General Coordinator of the Conference Pr. OULD HAMMOU Mustapha University of Boumerdes Dr.ROUAM Moussa : Pr. TURKMEN Mutlu University of Bayburt- Turkey The Organizing Committee Coordinator Organizes Pr. BELGHOUL Fathi University of Algiers 3 Dr.KOUASSEH Nadhir Member Pr.AHMED youcef University of Benha- Egypt Dr.GUERMAT Nouri Member Pr.CHIHA Fouad University of Constantine 2 Dr.DJEBBAR Abd el salem Member Pr.BENKARA Yacin University of Constantine 2 Dr.LAROUI Ilyes Member Pr. CHERIFI Ali University of Algiers 3 Pr.HANY Eldesouky University of South Valley - Egypt Pr.AHMED Sewilam Damietta University- Egypt Pr.MHIMDET Rachid CREPS.Constantine The International Virtual Conference Entitled: Dr.BOUBAKER Abdelkerim University of Menouba - Tunisia Dr. MERABET Messaoud University of Oum El Bouaghi Physical Preparation and Performance Dr. BENFADEL Fouad University of Oum El Bouaghi Dr. GASMI Abdelmalek University of Batna Analysis Technology in High Level Athletes Dr. LATRECHE Zoubir University of Oum El Bouaghi The Conference Secretariat: Dr. BOUNEB Chakeur University of Oum El Bouaghi April 10-11, 2021 Via google meet Dr.