Genetics and Major Depressive Disorder
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Effects of Serotonin in the Hippocampus: How Ssris and Multimodal Antidepressants Might Regulate Pyramidal Cell Function
CNS Spectrums (2016), 21, 143–161. © Cambridge University Press 2015. The online version of this article is published within an Open Access environment subject to the conditions of the Creative Commons Attribution-NonCommercial-ShareAlike licence <http://creativecommons.org/licenses/by-nc-sa/3.0/>. The written permission of Cambridge University Press must be obtained for commercial re-use. doi:10.1017/S1092852915000425 REVIEW ARTICLE Effects of serotonin in the hippocampus: how SSRIs and multimodal antidepressants might regulate pyramidal cell function Elena Dale,1* Alan L. Pehrson,1 Theepica Jeyarajah,1 Yan Li,1 Steven C. Leiser,1 Gennady Smagin,1 Christina K. Olsen,2 and Connie Sanchez1 1 Lundbeck Research USA, Paramus, New Jersey, USA 2 Lundbeck DK, Copenhagen-Valby, Denmark The hippocampus plays an important role in emotional and cognitive processing, and both of these domains are affected in patients with major depressive disorder (MDD). Extensive preclinical research and the notion that modulation of serotonin (5-HT) neurotransmission plays a key role in the therapeutic efficacy of selective serotonin reuptake inhibitors (SSRIs) support the view that 5-HT is important for hippocampal function in normal and disease- like conditions. The hippocampus is densely innervated by serotonergic fibers, and the majority of 5-HT receptor subtypes are expressed there. Furthermore, hippocampal cells often co-express multiple 5-HT receptor subtypes that can have either complementary or opposing effects on cell function, adding to the complexity of 5-HT neurotransmission. Here we review the current knowledge of how 5-HT, through its various receptor subtypes, modulates hippocampal output and the activity of hippocampal pyramidal cells in rodents. -

Stems for Nonproprietary Drug Names
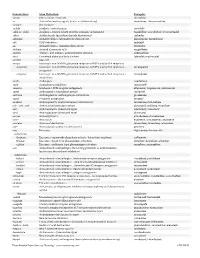
USAN STEM LIST STEM DEFINITION EXAMPLES -abine (see -arabine, -citabine) -ac anti-inflammatory agents (acetic acid derivatives) bromfenac dexpemedolac -acetam (see -racetam) -adol or analgesics (mixed opiate receptor agonists/ tazadolene -adol- antagonists) spiradolene levonantradol -adox antibacterials (quinoline dioxide derivatives) carbadox -afenone antiarrhythmics (propafenone derivatives) alprafenone diprafenonex -afil PDE5 inhibitors tadalafil -aj- antiarrhythmics (ajmaline derivatives) lorajmine -aldrate antacid aluminum salts magaldrate -algron alpha1 - and alpha2 - adrenoreceptor agonists dabuzalgron -alol combined alpha and beta blockers labetalol medroxalol -amidis antimyloidotics tafamidis -amivir (see -vir) -ampa ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) subgroup: -ampanel antagonists becampanel -ampator modulators forampator -anib angiogenesis inhibitors pegaptanib cediranib 1 subgroup: -siranib siRNA bevasiranib -andr- androgens nandrolone -anserin serotonin 5-HT2 receptor antagonists altanserin tropanserin adatanserin -antel anthelmintics (undefined group) carbantel subgroup: -quantel 2-deoxoparaherquamide A derivatives derquantel -antrone antineoplastics; anthraquinone derivatives pixantrone -apsel P-selectin antagonists torapsel -arabine antineoplastics (arabinofuranosyl derivatives) fazarabine fludarabine aril-, -aril, -aril- antiviral (arildone derivatives) pleconaril arildone fosarilate -arit antirheumatics (lobenzarit type) lobenzarit clobuzarit -arol anticoagulants (dicumarol type) dicumarol -
Downloaded from Survive Nursing | Survivenursing.Com V20110426
Generic Stem Stem Definition Examples -abine (see -arabine, -citabine) decitabine -ac Anti-inflammatory agents (acetic acid derivatives) bromfenac; dexpemedolac -acetam See -racetam -actide Synthetic corticotropins seractide -adol or -aldol- Analgesics (mixed opiate receptor agonists/ antagonists) tazadolene; spiradolene; levonantradol -adox Antibacterials (quinoline dioxide derivatives) carbadox -afenone Antiarrhythmics (propafenone derivatives) alprafenone; diprafenone -afil PDE5 inhibitors tadalafil -aj- Antiarrhythmics (ajmaline derivatives) lorajmine -aldrate Antacid aluminum salts magaldrate -algron Alpha1 - and alpha2 - adrenoreceptor agonists dabuzalgron -alol Combined alpha and beta blockers labetalol; medroxalol -amivir (see -vir) -ampa Ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) -ampanel Ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) ; becampanel antagonists -ampator Ionotropic non-NMDA glutamate receptors (AMPA and/or KA receptors) ; forampator modulators -andr- Androgens nandrolone -anib Angiogenesis inhibitors semaxanib -anserin Serotonin 5-HT2 receptor antagonists altanserin; tropanserin; adatanserin -antel Anthelmintics (undefined group) carbantel -antrone Antineoplastics; anthraquinone derivatives pixantrone -apsel P-selectin antagonists torapsel -arabine Antineoplastics (arabinofuranosyl derivatives) fazarabine; fludarabine aril-, -aril, -aril- Antiviral (arildone derivatives) pleconaril; arildone; fosarilate -arit Antirheumatics (lobenzarit type) lobenzarit; clobuzarit -arol -

United States Patent (10) Patent No.: US 7,368,477 B2 Gross Et Al
USOO7368477B2 (12) United States Patent (10) Patent No.: US 7,368,477 B2 Gross et al. (45) Date of Patent: May 6, 2008 (54) BENZOFURANYLALKANAMINE 6,419,958 B2 7/2002 Sherman et al. DERVATIVES AND USES THEREOF 6,444,708 B2 9/2002 Rudolph et al. 6,765,008 B1 7, 2004 Chen (75) Inventors: Jonathan Laird Gross, Cranbury, NJ 6,967.201 B1 1 1/2005 Briner et al. (US); Gary Paul Stack, Ambler, PA 7,045,545 B1 5, 2006 Briner et al. (US); Dahui Zhou, East Brunswick, NJ 2002/0183395 Al 12/2002 Argentieri et al. (US); Hong Gao, Belle Mead, NJ (US) 2004.0029949 A1 2/2004 Argentieri et al. 2004/0235856 Al 11/2004 McMurray et al. (73) Assignee: Wyeth, Madison, NJ (US) 2005, 0124692 A1 6/2005 Gross et al. 2005.01434.52 A1 6/2005 Gross et al. (*) Notice: Subject to any disclaimer, the term of this 2005/0261347 A1 11/2005 Gross et al. patent is extended or adjusted under 35 2006,008.9405 A1 4, 2006 Zhou U.S.C. 154(b) by 99 days. 2006/0111438 A1 5/2006 Gontcharov et al. 2006/0241172 A1 10, 2006 Zhou et al. (21) Appl. No.: 11/408,319 2006/0241176 A1 10, 2006 Stack et al. 2006/0246551 A1 11/2006 Stack et al. (22) Filed: Apr. 21, 2006 2006/0247276 A1 11/2006 Gross et al. 2006/0252825 A1 11/2006 Tadayon et al. (65) Prior Publication Data 2006/0258639 A1 11/2006 Logue et al. 2006/0258711 A1 11/2006 Rosenzweig-Lipson US 2006/O24727.6 A1 Nov. -

A Abacavir Abacavirum Abakaviiri Abagovomab Abagovomabum
A abacavir abacavirum abakaviiri abagovomab abagovomabum abagovomabi abamectin abamectinum abamektiini abametapir abametapirum abametapiiri abanoquil abanoquilum abanokiili abaperidone abaperidonum abaperidoni abarelix abarelixum abareliksi abatacept abataceptum abatasepti abciximab abciximabum absiksimabi abecarnil abecarnilum abekarniili abediterol abediterolum abediteroli abetimus abetimusum abetimuusi abexinostat abexinostatum abeksinostaatti abicipar pegol abiciparum pegolum abisipaaripegoli abiraterone abirateronum abirateroni abitesartan abitesartanum abitesartaani ablukast ablukastum ablukasti abrilumab abrilumabum abrilumabi abrineurin abrineurinum abrineuriini abunidazol abunidazolum abunidatsoli acadesine acadesinum akadesiini acamprosate acamprosatum akamprosaatti acarbose acarbosum akarboosi acebrochol acebrocholum asebrokoli aceburic acid acidum aceburicum asebuurihappo acebutolol acebutololum asebutololi acecainide acecainidum asekainidi acecarbromal acecarbromalum asekarbromaali aceclidine aceclidinum aseklidiini aceclofenac aceclofenacum aseklofenaakki acedapsone acedapsonum asedapsoni acediasulfone sodium acediasulfonum natricum asediasulfoninatrium acefluranol acefluranolum asefluranoli acefurtiamine acefurtiaminum asefurtiamiini acefylline clofibrol acefyllinum clofibrolum asefylliiniklofibroli acefylline piperazine acefyllinum piperazinum asefylliinipiperatsiini aceglatone aceglatonum aseglatoni aceglutamide aceglutamidum aseglutamidi acemannan acemannanum asemannaani acemetacin acemetacinum asemetasiini aceneuramic -

PHARMACEUTICAL APPENDIX to the TARIFF SCHEDULE 2 Table 1
Harmonized Tariff Schedule of the United States (2010) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2010) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. -

(12) Patent Application Publication (10) Pub. No.: US 2010/0184806 A1 Barlow Et Al
US 20100184806A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2010/0184806 A1 Barlow et al. (43) Pub. Date: Jul. 22, 2010 (54) MODULATION OF NEUROGENESIS BY PPAR (60) Provisional application No. 60/826,206, filed on Sep. AGENTS 19, 2006. (75) Inventors: Carrolee Barlow, Del Mar, CA (US); Todd Carter, San Diego, CA Publication Classification (US); Andrew Morse, San Diego, (51) Int. Cl. CA (US); Kai Treuner, San Diego, A6II 3/4433 (2006.01) CA (US); Kym Lorrain, San A6II 3/4439 (2006.01) Diego, CA (US) A6IP 25/00 (2006.01) A6IP 25/28 (2006.01) Correspondence Address: A6IP 25/18 (2006.01) SUGHRUE MION, PLLC A6IP 25/22 (2006.01) 2100 PENNSYLVANIA AVENUE, N.W., SUITE 8OO (52) U.S. Cl. ......................................... 514/337; 514/342 WASHINGTON, DC 20037 (US) (57) ABSTRACT (73) Assignee: BrainCells, Inc., San Diego, CA (US) The instant disclosure describes methods for treating diseases and conditions of the central and peripheral nervous system (21) Appl. No.: 12/690,915 including by stimulating or increasing neurogenesis, neuro proliferation, and/or neurodifferentiation. The disclosure (22) Filed: Jan. 20, 2010 includes compositions and methods based on use of a peroxi some proliferator-activated receptor (PPAR) agent, option Related U.S. Application Data ally in combination with one or more neurogenic agents, to (63) Continuation-in-part of application No. 1 1/857,221, stimulate or increase a neurogenic response and/or to treat a filed on Sep. 18, 2007. nervous system disease or disorder. Patent Application Publication Jul. 22, 2010 Sheet 1 of 9 US 2010/O184806 A1 Figure 1: Human Neurogenesis Assay Ciprofibrate Neuronal Differentiation (TUJ1) 100 8090 Ciprofibrates 10-8.5 10-8.0 10-7.5 10-7.0 10-6.5 10-6.0 10-5.5 10-5.0 10-4.5 Conc(M) Patent Application Publication Jul. -

Pharmaceutical Appendix to the Tariff Schedule 2
Harmonized Tariff Schedule of the United States (2015) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2015) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. -
PHARMACEUTICAL APPENDIX to the HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2011) (Rev
Harmonized Tariff Schedule of the United States (2011) (Rev. 1) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2011) (Rev. 1) Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. ABACAVIR 136470-78-5 ACEVALTRATE 25161-41-5 ABAFUNGIN 129639-79-8 ACEXAMIC ACID 57-08-9 ABAGOVOMAB 792921-10-9 ACICLOVIR 59277-89-3 ABAMECTIN 65195-55-3 ACIFRAN 72420-38-3 ABANOQUIL 90402-40-7 ACIPIMOX 51037-30-0 ABAPERIDONE 183849-43-6 ACITAZANOLAST 114607-46-4 ABARELIX 183552-38-7 ACITEMATE 101197-99-3 ABATACEPT 332348-12-6 ACITRETIN 55079-83-9 ABCIXIMAB 143653-53-6 ACIVICIN 42228-92-2 ABECARNIL 111841-85-1 ACLANTATE 39633-62-0 ABETIMUS 167362-48-3 ACLARUBICIN 57576-44-0 ABIRATERONE 154229-19-3 ACLATONIUM NAPADISILATE 55077-30-0 ABITESARTAN 137882-98-5 ACLIDINIUM BROMIDE 320345-99-1 ABLUKAST 96566-25-5 ACODAZOLE 79152-85-5 ABRINEURIN 178535-93-8 ACOLBIFENE 182167-02-8 ABUNIDAZOLE 91017-58-2 ACONIAZIDE 13410-86-1 ACADESINE 2627-69-2 ACOTIAMIDE 185106-16-5 -
Chemical Structure-Related Drug-Like Criteria of Global Approved Drugs
Molecules 2016, 21, 75; doi:10.3390/molecules21010075 S1 of S110 Supplementary Materials: Chemical Structure-Related Drug-Like Criteria of Global Approved Drugs Fei Mao 1, Wei Ni 1, Xiang Xu 1, Hui Wang 1, Jing Wang 1, Min Ji 1 and Jian Li * Table S1. Common names, indications, CAS Registry Numbers and molecular formulas of 6891 approved drugs. Common Name Indication CAS Number Oral Molecular Formula Abacavir Antiviral 136470-78-5 Y C14H18N6O Abafungin Antifungal 129639-79-8 C21H22N4OS Abamectin Component B1a Anthelminithic 65195-55-3 C48H72O14 Abamectin Component B1b Anthelminithic 65195-56-4 C47H70O14 Abanoquil Adrenergic 90402-40-7 C22H25N3O4 Abaperidone Antipsychotic 183849-43-6 C25H25FN2O5 Abecarnil Anxiolytic 111841-85-1 Y C24H24N2O4 Abiraterone Antineoplastic 154229-19-3 Y C24H31NO Abitesartan Antihypertensive 137882-98-5 C26H31N5O3 Ablukast Bronchodilator 96566-25-5 C28H34O8 Abunidazole Antifungal 91017-58-2 C15H19N3O4 Acadesine Cardiotonic 2627-69-2 Y C9H14N4O5 Acamprosate Alcohol Deterrant 77337-76-9 Y C5H11NO4S Acaprazine Nootropic 55485-20-6 Y C15H21Cl2N3O Acarbose Antidiabetic 56180-94-0 Y C25H43NO18 Acebrochol Steroid 514-50-1 C29H48Br2O2 Acebutolol Antihypertensive 37517-30-9 Y C18H28N2O4 Acecainide Antiarrhythmic 32795-44-1 Y C15H23N3O2 Acecarbromal Sedative 77-66-7 Y C9H15BrN2O3 Aceclidine Cholinergic 827-61-2 C9H15NO2 Aceclofenac Antiinflammatory 89796-99-6 Y C16H13Cl2NO4 Acedapsone Antibiotic 77-46-3 C16H16N2O4S Acediasulfone Sodium Antibiotic 80-03-5 C14H14N2O4S Acedoben Nootropic 556-08-1 C9H9NO3 Acefluranol Steroid -

Cho-Ming Loi, Dennis A. Smith and Deepak Dalvie Pharmacokinetics
Drug Metabolism and Disposition Which Metabolites Circulate? Cho-Ming Loi, Dennis A. Smith and Deepak Dalvie Pharmacokinetics, Dynamics, and Metabolism, Pfizer Worldwide Research, San Diego, CA 92121 (C-M L and DD) and Honorary Professor, Department of Chemistry, University of Capetown, Capetown, South Africa, Visiting Professor, Institute of Translational Medicine, University of Liverpool, UK (DAS). 1 SupplTable 1: Structures of drugs and their amine metabolites Drugs Metabolites O I H N O O I Amiodarone N-Desethylamiodarone Didesethylamiodarone N Amitriptyline Nortriptyline Atomoxetine Desmethylatomoxetine Azelastine Desmethylazelastine Cl O NH N O N N N O Azimilide Desmethylazilimide 1-Pyrimidinylpiperazine Buspirone 2 Citalopram Desmethylcitalopram Didesmethylcitalopram Clobazam Norclobazam Clomipramine Desmethylclomipramine Clozapine Desmethylclozapine H2N N N H N N O S N O O H Desisopropyldelavirdine Delavirdine Diazepam Desmethyldiazepam 3 Diltiazem Desmethyldiltiazem Dothiepin Desmethyldothiepin O N H Doxepin Desmethyldoxepin Enzalutamide Desmethylenzalutamide Elzasonan Desmethyl elzasonan Fenfluramine Desethylfenfluramine N O N Norfentanyl Fentanyl Fluoxetine Norfluoxetine 4 Halofantrine Desbutylhalofantrine N N N N N H NNH O Imatinib Desmethylimatinib Imipramine Desipramine Itraconazole Dealkylitraconazole Ivabradine Desmethylivabradine Desloratadine Loratadine Loxapine Desmethylloxapine 5 Lumefantrine Desbutyllumefantrine Mianserin Desmethylmianserin Mitrazapine Desmethylmirtazapine m-Chlorophenylpiperazine Nefazodone Olanzapine -

Customs Tariff - Schedule
CUSTOMS TARIFF - SCHEDULE 99 - i Chapter 99 SPECIAL CLASSIFICATION PROVISIONS - COMMERCIAL Notes. 1. The provisions of this Chapter are not subject to the rule of specificity in General Interpretative Rule 3 (a). 2. Goods which may be classified under the provisions of Chapter 99, if also eligible for classification under the provisions of Chapter 98, shall be classified in Chapter 98. 3. Goods may be classified under a tariff item in this Chapter and be entitled to the Most-Favoured-Nation Tariff or a preferential tariff rate of customs duty under this Chapter that applies to those goods according to the tariff treatment applicable to their country of origin only after classification under a tariff item in Chapters 1 to 97 has been determined and the conditions of any Chapter 99 provision and any applicable regulations or orders in relation thereto have been met. 4. The words and expressions used in this Chapter have the same meaning as in Chapters 1 to 97. Issued January 1, 2018 99 - 1 CUSTOMS TARIFF - SCHEDULE Tariff Unit of MFN Applicable SS Description of Goods Item Meas. Tariff Preferential Tariffs 9901.00.00 Articles and materials for use in the manufacture or repair of the Free CCCT, LDCT, GPT, UST, following to be employed in commercial fishing or the commercial MT, MUST, CIAT, CT, harvesting of marine plants: CRT, IT, NT, SLT, PT, COLT, JT, PAT, HNT, Artificial bait; KRT, CEUT, UAT: Free Carapace measures; Cordage, fishing lines (including marlines), rope and twine, of a circumference not exceeding 38 mm; Devices for keeping nets open; Fish hooks; Fishing nets and netting; Jiggers; Line floats; Lobster traps; Lures; Marker buoys of any material excluding wood; Net floats; Scallop drag nets; Spat collectors and collector holders; Swivels.