Supplemental Tables
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Materials
SUPPLEMENTARY MATERIALS Table S1. Chemical characteristics of the two digestate forms (SD and WD). Values quoted are expressed as % of air-dry digestate (means followed by standard error in brackets). References for the employed methods used for determination of each chemical characteristic is also reported. SD WD Reference Org C % 44.4 (0.33) 1.1 (0.01) [81] Tot N % 1.4 (0.01) 0.4 (0.01) [82] C/N 31.4 (0.17) 3.1 (0.04) NH4-N % n.d 0.2 (0.00) [83] K % 1.7 (0.00) n.d. [84] P % 0.9 (0.01) n.d. [84] S % 0.23 (0.02) n.d. [85] SD = solid digestate; WD = whole digestate. Table S2. Soil physical and chemical characteristics at the beginning of trial (t0) (means from 9 observations followed by standard errors in brackets). Clay (%) 41.9 (1.22) Silt (%) 47.8 (2.13) Moisture (%) 24.46 (1.24) Bulk density (g cm-3) 1.39 (0.04) pH 8.3 (0) CaCO3 (%) 11.4 (0.7) TOC (g kg-1) 12.8 (0.3) TN (g kg-1) 1.4 (0) C/N 9.4 (0.3) CEC (cmol(+) kg-1) 21.0 (0.7) Exchangeable Bases (mg kg-1) K 278.7 (8.0) Na 22.0 (3.0) Mg 201.7 (25.6) Ca 3718.6 (176.3) Available Microelements (mg kg-1) Cu 28.0 (5.0) Zn 1.7 (0.2) Fe 15.4 (0.5) Mn 16.3 (0.5) TOC = total organic C; TN = total N; CEC = cation exchange capacity Table S3. -

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

Effect of Ph and Temperature on Microbial Community Structure And
Calicioglu et al. Biotechnol Biofuels (2018) 11:275 https://doi.org/10.1186/s13068-018-1278-6 Biotechnology for Biofuels RESEARCH Open Access Efect of pH and temperature on microbial community structure and carboxylic acid yield during the acidogenic digestion of duckweed Ozgul Calicioglu1, Michael J. Shreve1, Tom L. Richard2 and Rachel A. Brennan1* Abstract Background: Duckweeds (Lemnaceae) are efcient aquatic plants for wastewater treatment due to their high nutri- ent-uptake capabilities and resilience to severe environmental conditions. Combined with their rapid growth rates, high starch, and low lignin contents, duckweeds have also gained popularity as a biofuel feedstock for thermochemi- cal conversion and alcohol fermentation. However, studies on the acidogenic anaerobic digestion of duckweed into carboxylic acids, another group of chemicals which are precursors of higher-value chemicals and biofuels, are lacking. In this study, a series of laboratory batch experiments were performed to determine the favorable operating condi- tions (i.e., temperature and pH) to maximize carboxylic acid production from wastewater-derived duckweed during acidogenic digestion. Batch reactors with 25 g/l solid loading were operated anaerobically for 21 days under meso- philic (35 °C) or thermophilic (55 °C) conditions at an acidic (5.3) or basic (9.2) pH. At the conclusion of the experiment, the dominant microbial communities under various operating conditions were assessed using high-throughput sequencing. Results: The highest duckweed–carboxylic acid conversion of 388 28 mg acetic acid equivalent per gram volatile solids was observed under mesophilic and basic conditions, with an± average production rate of 0.59 g/l/day. This result is comparable to those reported for acidogenic digestion of other organics such as food waste. -

Supporting Information
Supporting Information Lozupone et al. 10.1073/pnas.0807339105 SI Methods nococcus, and Eubacterium grouped with members of other Determining the Environmental Distribution of Sequenced Genomes. named genera with high bootstrap support (Fig. 1A). One To obtain information on the lifestyle of the isolate and its reported member of the Bacteroidetes (Bacteroides capillosus) source, we looked at descriptive information from NCBI grouped firmly within the Firmicutes. This taxonomic error was (www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and other related not surprising because gut isolates have often been classified as publications. We also determined which 16S rRNA-based envi- Bacteroides based on an obligate anaerobe, Gram-negative, ronmental surveys of microbial assemblages deposited near- nonsporulating phenotype alone (6, 7). A more recent 16S identical sequences in GenBank. We first downloaded the gbenv rRNA-based analysis of the genus Clostridium defined phylo- files from the NCBI ftp site on December 31, 2007, and used genetically related clusters (4, 5), and these designations were them to create a BLAST database. These files contain GenBank supported in our phylogenetic analysis of the Clostridium species in the HGMI pipeline. We thus designated these Clostridium records for the ENV database, a component of the nonredun- species, along with the species from other named genera that dant nucleotide database (nt) where 16S rRNA environmental cluster with them in bootstrap supported nodes, as being within survey data are deposited. GenBank records for hits with Ͼ98% these clusters. sequence identity over 400 bp to the 16S rRNA sequence of each of the 67 genomes were parsed to get a list of study titles Annotation of GTs and GHs. -

Bacterial Profiles and Antibiogrants of the Bacteria Isolated of the Exposed Pulps of Dog and Cheetah Canine Teeth
Bacterial profiles and antibiogrants of the bacteria isolated of the exposed pulps of dog and cheetah canine teeth A dissertation submitted to the Faculty of Veterinary Science, University of Pretoria. In partial fulfillment of the requirements for the degree Master of Science (Veterinary Science) Promoter: Dr. Gerhard Steenkamp Co-promoter: Ms. Anna-Mari Bosman Department of Companion Animal Clinical Studies Faculty of Veterinary Science University of Pretoria Pretoria (January 2012) J.C. Almansa Ruiz © University of Pretoria Declaration I declare that the dissertation that I hereby submit for the Masters of Science degree in Veterinary Science at the University of Pretoria has not previously been submitted by me for degree purposes at any other university. J.C. Almansa Ruiz ii Dedications To one of the most amazing hunters of the African bush, the Cheetah, that has made me dream since I was a child, and the closest I had been to one, before starting this project was in National Geographic documentaries. I wish all of them a better future in which their habitat will be more respected. To all conservationists, especially to Carla Conradie and Dave Houghton, for spending their lives saving these animals which are suffering from the consequences of the encroachment of human beings into their territory. Some, such as George Adamson, the lion conservationist, even lost their lives in this mission. To the conservationist, Lawrence Anthony, for risking his life in a suicide mission to save the animals in the Baghdad Zoo, when the conflict in Iraq exploded. To my mother and father Jose Maria and Rosa. -

Significance of Donor Human Milk
fmicb-09-01376 June 26, 2018 Time: 17:31 # 1 ORIGINAL RESEARCH published: 27 June 2018 doi: 10.3389/fmicb.2018.01376 Preterm Gut Microbiome Depending on Feeding Type: Significance of Donor Human Milk Anna Parra-Llorca1, María Gormaz1,2, Cristina Alcántara3, María Cernada1,2, Antonio Nuñez-Ramiro1,2, Máximo Vento1,2*† and Maria C. Collado3*† 1 Neonatal Research Group, Health Research Institute La Fe, University and Polytechnic Hospital La Fe, Valencia, Spain, 2 Division of Neonatology, University and Polytechnic Hospital La Fe, Valencia, Spain, 3 Department of Biotechnology, Institute of Agrochemistry and Food Technology, Spanish National Research Council, Valencia, Spain Preterm microbial colonization is affected by gestational age, antibiotic treatment, type of birth, but also by type of feeding. Breast milk has been acknowledged as the gold standard for human nutrition. In preterm infants breast milk has been associated with improved growth and cognitive development and a reduced risk of necrotizing enterocolitis and late onset sepsis. In the absence of their mother’s own milk (MOM), pasteurized donor human milk (DHM) could be the best available alternative due to its similarity to the former. However, little is known about the effect of DHM upon preterm Edited by: Sandra Torriani, microbiota and potential biological implications. Our objective was to determine the University of Verona, Italy impact of DHM upon preterm gut microbiota admitted in a referral neonatal intensive Reviewed by: care unit (NICU). A prospective observational cohort study in NICU of 69 neonates Carlotta De Filippo, <32 weeks of gestation and with a birth weight ≤1,500 g was conducted. -

Production of Bio-Hydrogen from Dairy Wastewater Using Pretreated Landfill
Journal of Bioscience and Bioengineering VOL. 127 No. 2, 150e159, 2019 www.elsevier.com/locate/jbiosc Production of bio-hydrogen from dairy wastewater using pretreated landfill leachate sludge as an inoculum Yee Meng Wong,1,2 Pau Loke Show,3 Ta Yeong Wu,4 Hui Yi Leong,3 Shaliza Ibrahim,5 and Joon Ching Juan1,2,* School of Science, Monash University Malaysia, Jalan Lagoon Selatan, Bandar Sunway, 46150 Selangor Darul Ehsan, Malaysia,1 Nanotechnology & Catalysis Research Centre (NANOCAT), University of Malaya, 50603 Kuala Lumpur, Malaysia,2 Bioseparation Research Group, Department of Chemical and Environmental Engineering, Faculty of Engineering, University of Nottingham Malaysia Campus, Jalan Broga, 43500 Semenyih, Selangor Darul Ehsan, Malaysia,3 Chemical Engineering Discipline, School of Engineering, Monash University, Jalan Lagoon Selatan, Bandar Sunway, 46150 Selangor Darul Ehsan, Malaysia,4 and Department of Civil Engineering, Faculty of Engineering, University of Malaya, 50603 Kuala Lumpur, Malaysia5 Received 26 February 2018; accepted 15 July 2018 Available online 15 September 2018 Bio-hydrogen production from wastewater using sludge as inoculum is a sustainable approach for energy production. This study investigated the influence of initial pH and temperature on bio-hydrogen production from dairy wastewater using pretreated landfill leachate sludge (LLS) as an inoculum. The maximum yield of 113.2 ± 2.9 mmol H2/g chemical oxygen demand (COD) (12.8 ± 0.3 mmol H2/g carbohydrates) was obtained at initial pH 6 and 37 C. The main products of volatile fatty acids were acetate and butyrate with the ratio of acetate:butyrate was 0.4. At optimum condition, Gibb’s free energy was estimated at L40 kJ/mol, whereas the activation enthalpy and entropy were 65 kJ/mol and 0.128 kJ/mol/ l, respectively. -

Clostridium Amazonitimonense, Clostridium Me
ORIGINAL ARTICLE Taxonogenomic description of four new Clostridium species isolated from human gut: ‘Clostridium amazonitimonense’, ‘Clostridium merdae’, ‘Clostridium massilidielmoense’ and ‘Clostridium nigeriense’ M. T. Alou1, S. Ndongo1, L. Frégère1, N. Labas1, C. Andrieu1, M. Richez1, C. Couderc1, J.-P. Baudoin1, J. Abrahão2, S. Brah3, A. Diallo1,4, C. Sokhna1,4, N. Cassir1, B. La Scola1, F. Cadoret1 and D. Raoult1,5 1) Aix-Marseille Université, Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes, UM63, CNRS 7278, IRD 198, INSERM 1095, Marseille, France, 2) Laboratório de Vírus, Departamento de Microbiologia, Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil, 3) Hopital National de Niamey, BP 247, Niamey, Niger, 4) Campus Commun UCAD-IRD of Hann, Route des pères Maristes, Hann Maristes, BP 1386, CP 18524, Dakar, Senegal and 5) Special Infectious Agents Unit, King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia Abstract Culturomics investigates microbial diversity of the human microbiome by combining diversified culture conditions, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and 16S rRNA gene identification. The present study allowed identification of four putative new Clostridium sensu stricto species: ‘Clostridium amazonitimonense’ strain LF2T, ‘Clostridium massilidielmoense’ strain MT26T, ‘Clostridium nigeriense’ strain Marseille-P2414T and ‘Clostridium merdae’ strain Marseille-P2953T, which we describe using the concept of taxonogenomics. We describe the main characteristics of each bacterium and present their complete genome sequence and annotation. © 2017 Published by Elsevier Ltd. Keywords: ‘Clostridium amazonitimonense’, ‘Clostridium massilidielmoense’, ‘Clostridium merdae’, ‘Clostridium nigeriense’, culturomics, emerging bacteria, human microbiota, taxonogenomics Original Submission: 18 August 2017; Revised Submission: 9 November 2017; Accepted: 16 November 2017 Article published online: 22 November 2017 intestine [1,4–6]. -

Serpentinicella Alkaliphila Gen. Nov., Sp. Nov., a Novel
International Journal of Systematic and Evolutionary Microbiology (2016), 66, 4464–4470 DOI 10.1099/ijsem.0.001375 Serpentinicella alkaliphila gen. nov., sp. nov., a novel alkaliphilic anaerobic bacterium isolated from the serpentinite-hosted Prony hydrothermal field, New Caledonia. Nan Mei,1 Anne Postec,1 Gael Erauso,1 Manon Joseph,1 Bernard Pelletier,2 Claude Payri,2 Bernard Ollivier1 and Marianne Quem eneur 1 Correspondence 1Aix Marseille Univ, Universite de Toulon, CNRS, IRD, MIO, Marseille, France Marianne Quemeneur 2Centre IRD de Noumea, 101 Promenade Roger Laroque, BP A5 - 98848 Noumea cedex, , New [email protected] Caledonia A novel anaerobic, alkaliphilic, Gram-stain-positive, spore-forming bacterium was isolated from a carbonaceous hydrothermal chimney in Prony Bay, New Caledonia. This bacterium, designated strain 3bT, grew at temperatures from 30 to 43 C (optimum 37 C) and at pH between 7.8 and 10.1 (optimum 9.5). Added NaCl was not required for growth (optimum 0–0.2 %, w/v), but was tolerated at up to 4 %. Yeast extract was required for growth. Strain 3bT utilized crotonate, lactate and pyruvate, but not sugars. Crotonate was dismutated to acetate and butyrate. Lactate was disproportionated to acetate and propionate. Pyruvate was degraded to acetate plus trace amounts of hydrogen. Growth on lactate was improved by the addition of fumarate, which was used as an electron acceptor and converted to succinate. Sulfate, thiosulfate, elemental sulfur, sulfite, nitrate, nitrite, FeCl3, Fe(III)-citrate, Fe(III)-EDTA, chromate, arsenate, selenate and DMSO were not used as terminal electron acceptors. The G+C content of the genomic DNA was 33.2 mol%. -

United States Patent (10) Patent No.: US 7820,184 B2 Stritzker Et Al
USOO782O184B2 (12) United States Patent (10) Patent No.: US 7820,184 B2 Stritzker et al. (45) Date of Patent: Oct. 26, 2010 (54) METHODS AND COMPOSITIONS FOR 5,833,975 A 1 1/1998 Paoletti et al. ............. 424.93.2 DETECTION OF MICROORGANISMS AND 5,976,796. A 1 1/1999 Szalay et al. ................... 435/6 SirNRTREATMENT OF DISEASES AND 6,025,155 A 2/2000 Hadlaczky et al. ......... 435/69.1 6,045,802 A 4/2000 Schlom et al. ........... 424,199.1 (75) Inventors: Jochen Harald Stritzker, Kissing (DE); 6,077,697 A 6/2000 Hadlaczky et al. 435/1723 Phil Hill, West Bridgford (GB); Aladar 6,080,849 A 6/2000 Bermudes et al. .......... 536,23.7 A. Szalay, Highland, CA (US); Yong A. 6,093,700 A 7/2000 Mastrangelo et al. ......... 514,44 Yu, San Diego, CA (US) 6,099,848. A 8/2000 Frankel et al. ........... 424,246.1 6,106,826 A 8/2000 Brandt et al. .............. 424.93.2 (73) Assignee: stylus Corporation, San Diego, CA 6, 190,657 B1 2/2001 Pawelek et al. ............ 424,931 6,217,847 B1 4/2001 Contaget al. ................ 4249.1 (*) Notice: Subject to any disclaimer, the term of this 6,232,523 B1 5/2001 Tan et al. ...................... 800, 10 patent is extended or adjusted under 35 6,235,967 B1 5/2001 Tan et al. ...................... 800, 10 U.S.C. 154(b) by 362 days. 6,235,968 B1 5/2001 Tan et al. ...................... 800, 10 6,251,384 B1 6/2001 Tan et al. -
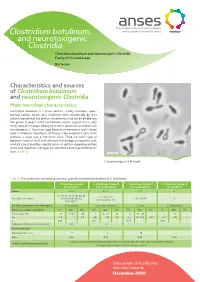
Clostridium Botulinum, and Neurotoxigenic Clostridia Clostridium Botulinum and Neurotoxigenic Clostridia Family of Clostridiaceae Bacterium
Clostridium botulinum, and neurotoxigenic Clostridia Clostridium botulinum and neurotoxigenic Clostridia Family of Clostridiaceae Bacterium Characteristics and sources of Clostridium botulinum and neurotoxigenic Clostridia Main microbial characteristics Clostridium botulinum is a Gram-positive, strictly anaerobic, spore- forming bacillus. Strains of C. botulinum differ considerably by their cultural, biochemical and genetic characteristics and can be divided into four groups (Groups I to IV). Furthermore, certain atypical strains, only rarely isolated in Europe, belonging to other species of Clostridium, are neurotoxigenic: C. butyricum (type E botulinum neurotoxin) and C. baratii (type F botulinum neurotoxin). With only a few exceptions, each strain produces a single type of botulinum toxin. There are seven types of botulinum toxin (A to G) with different immunological properties, each of which is neutralised by a specific serum. In addition, depending on their amino acid sequences, sub-types are identified in each type of botulinum toxin (Table 1). C. botulinum type A. © M. Popoff Table 1. Characteristics concerning survival, growth and toxin production of C. botulinum C. botulinum Group I C. botulinum Group II C. botulinum Group III C. botulinum Group IV (proteolytic) (non-proteolytic) (non-proteolytic) (proteolytic) Toxins A, B, F B, E, F C, D G A1, A2, A3, A4, A5, B1, B2, B3, E1, E2, E3, E6, Sous-types de toxines bivalent B (Ba, Bf, Ab), C, D, C/D, D/C G non-proteolytic B, F proteolytic F Bactéries apparentées non toxinogènes C. sporogenes Pas de nom d’espèces C. novyi C. subterminale Croissance cellules végétatives Min. Opt. Max. Min. Opt. Max. Min. Opt. Max. Min. -

Draft Genome Sequence of Dethiobacter Alkaliphilus Strain AHT1T, a Gram-Positive Sulfidogenic Polyextremophile Emily Denise Melton1, Dimitry Y
Melton et al. Standards in Genomic Sciences (2017) 12:57 DOI 10.1186/s40793-017-0268-9 EXTENDED GENOME REPORT Open Access Draft genome sequence of Dethiobacter alkaliphilus strain AHT1T, a gram-positive sulfidogenic polyextremophile Emily Denise Melton1, Dimitry Y. Sorokin2,3, Lex Overmars1, Alla L. Lapidus4, Manoj Pillay6, Natalia Ivanova5, Tijana Glavina del Rio5, Nikos C. Kyrpides5,6,7, Tanja Woyke5 and Gerard Muyzer1* Abstract Dethiobacter alkaliphilus strain AHT1T is an anaerobic, sulfidogenic, moderately salt-tolerant alkaliphilic chemolithotroph isolated from hypersaline soda lake sediments in northeastern Mongolia. It is a Gram-positive bacterium with low GC content, within the phylum Firmicutes. Here we report its draft genome sequence, which consists of 34 contigs with a total sequence length of 3.12 Mbp. D. alkaliphilus strain AHT1T was sequenced by the Joint Genome Institute (JGI) as part of the Community Science Program due to its relevance to bioremediation and biotechnological applications. Keywords: Extreme environment, Soda lake, Sediment, Haloalkaliphilic, Gram-positive, Firmicutes Introduction genome of a haloalkaliphilic Gram-positive sulfur dispro- Soda lakes are formed in environments where high rates portionator within the phylum Firmicutes: Dethiobacter of evaporation lead to the accumulation of soluble carbon- alkaliphilus AHT1T. ate salts due to the lack of dissolved divalent cations. Con- sequently, soda lakes are defined by their high salinity and Organism information stable highly alkaline pH conditions, making them dually Classification and features extreme environments. Soda lakes occur throughout the The haloalkaliphilic anaerobe D. alkaliphilus AHT1T American, European, African, Asian and Australian conti- was isolated from hypersaline soda lake sediments in nents and host a wide variety of Archaea and Bacteria, northeastern Mongolia [10].