Based on Chloroplast Trnl-Trnf Sequences
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

20. Tribe DESMODIEAE 116. TRIFIDACANTHUS Merrill, Philipp
20. Tribe DESMODIEAE 山蚂蝗族 shan ma huang zu Huang Puhua (黄普华 Huang Pu-hwa); Hiroyoshi Ohashi, Yu Iokawa, Tomoyuki Nemoto Herbs or shrubs, rarely trees or twining. Leaves pinnately 3(–9)-foliolate or 1-foliolate; stipules mostly striate; stipels present or sometimes absent. Flowers in terminal or axillary racemes or arranged into a panicle, rarely an umbel or fascicle. Calyx 4- or 5- toothed or 2-lipped. Wings equal to or exceeding keel and often adherent to it near base. Vexillary filament free or connate with others, sometimes forming a closed tube; anthers uniform. Legumes transversely jointed, sometimes of only 1 article, or rarely 2- valved. Seeds without a strophiole, rarely arillate. About 30 genera and 520–530 species: distributed in tropical, subtropical, and warm-temperate regions, but extending into the cool-temperate and sub-boreal regions of E Asia and North America; 18 genera and 139 species (42 endemic, four introduced) in China. 1a. Stipels absent, rarely present; legumes 1-jointed, 1-seeded, not glochidiate. 2a. Lateral veins of leaflets strict, extending to margin; stipules large, ovate, strongly ribbed ........................... 133. Kummerowia 2b. Lateral veins of leaflets arcuate, not reaching to margin; stipules small, subulate. 3a. Bracts 1-flowered, usually caducous; pedicels articulate below calyx; keel falcate, acute ................... 131. Campylotropis 3b. Bracts 2-flowered, persistent; pedicels not articulate; keel strict, obtuse ..................................................... 132. Lespedeza 1b. Stipels present; legumes usually glochidiate, 2- to several jointed, rarely 1-jointed, 1-seeded. 4a. Branch nodes with 3-fid, hard spines; leaves 1-foliolate ............................................................................... 116. Trifidacanthus 4b. Branch nodes without 3-fid, hard spines; leaves 3(–9)-foliolate, rarely 1-foliolate. -

Dendrolobium Triangulare, Desmodium Gangeticum, Desmodium Heterocarpon, and Tadehagi Triquetrum)
Mongabay.com Open Access Journal - Tropical Conservation Science Vol.2(1):52-69, 2009 Research Article Genetic relationships among accessions of four species of Desmodium and allied genera (Dendrolobium triangulare, Desmodium gangeticum, Desmodium heterocarpon, and Tadehagi triquetrum) Bettina Heider1*, Elke Fischer1, Tanja Berndl1, and Rainer Schultze-Kraft1,2 1Institute for Plant Production and Agroecology in the Tropics and Subtropics, University of Hohenheim, Garbenstrasse 13, D-70599 Stuttgart, Germany *E-mail: [email protected] 2 Centro Internacional de Agricultura Tropical (CIAT), A.A. 6713, Cali, Colombia Abstract Random amplified polymorphic DNA markers (RAPD) were used to assess the genetic relatedness among accessions of four species of Desmodium and allied genera (Dendrolobium triangulare, Desmodium gangeticum, Desmodium heterocarpon ssp. heterocarpon, and Tadehagi triquetrum) originating from Northeast Vietnam. Since information on the genetic diversity of these species is deficient, the creation of baseline data is an important means for the development of more sustainable and cost-efficient conservation approaches which eventually result in more comprehensive ex situ germplasm collections. The species analyzed are native to tropical and subtropical Asia, Australia, and Oceania and possess a potential as forage and/or medicinal plants. Moderate levels of inter-accession diversity represented by 37.5% and 33.3% of polymorphic fragments (P%) and average Jaccard’s similarity coefficients (JSCs) of 0.60 and 0.64 were found in D. heterocarpon and T. triquetrum, respectively, while moderate to high levels were detected in D. triangulare (P% = 52.9 and JSC = 0.61) and D. gangeticum (P% = 34.5 and JSC = 0.49). Mantel tests failed to reveal a correlation between geographic and genetic distances. -

Evolution of Angiosperm Pollen. 7. Nitrogen-Fixing Clade1
Evolution of Angiosperm Pollen. 7. Nitrogen-Fixing Clade1 Authors: Jiang, Wei, He, Hua-Jie, Lu, Lu, Burgess, Kevin S., Wang, Hong, et. al. Source: Annals of the Missouri Botanical Garden, 104(2) : 171-229 Published By: Missouri Botanical Garden Press URL: https://doi.org/10.3417/2019337 BioOne Complete (complete.BioOne.org) is a full-text database of 200 subscribed and open-access titles in the biological, ecological, and environmental sciences published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Complete website, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/terms-of-use. Usage of BioOne Complete content is strictly limited to personal, educational, and non - commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Downloaded From: https://bioone.org/journals/Annals-of-the-Missouri-Botanical-Garden on 01 Apr 2020 Terms of Use: https://bioone.org/terms-of-use Access provided by Kunming Institute of Botany, CAS Volume 104 Annals Number 2 of the R 2019 Missouri Botanical Garden EVOLUTION OF ANGIOSPERM Wei Jiang,2,3,7 Hua-Jie He,4,7 Lu Lu,2,5 POLLEN. 7. NITROGEN-FIXING Kevin S. Burgess,6 Hong Wang,2* and 2,4 CLADE1 De-Zhu Li * ABSTRACT Nitrogen-fixing symbiosis in root nodules is known in only 10 families, which are distributed among a clade of four orders and delimited as the nitrogen-fixing clade. -

Additions to Paleotropical Bruchidius Associated with Desmodieae (Coleoptera: Chrysomelidae: Bruchinae)
Genus Vol. 25(3): 425-432 Wrocław, 30 IX 2014 Additions to paleotropical Bruchidius associated with Desmodieae (Coleoptera: Chrysomelidae: Bruchinae) ALEX DELOBEL Muséum national d’Histoire naturelle, 45 rue Buffon, 75005 Paris, e-mail: [email protected] ABSTRACT. Three species of Bruchinae are reported for the first time from Vietnam as feeding in seeds of members of leguminous tribe Desmodieae (Fabaceae - Faboideae). Two of them are new to science and described in genus Bruchidius: B. hoangi and B. madaguiensis. A redescription of B. minutissimus (Motschulsky) is also provided. Key words: entomology, taxonomy, Coleoptera, Bruchinae, new species, Desmodieae, Alhagi, Dendrolobium, Desmodium. InTroDuCTIon Desmodieae are a leguminous tribe of Faboideae (or Papilionoideae) with nume- rous members in tropical regions; it is composed of 27 genera, of which Desmodium and Lespedeza are the largest. About 190 Desmodieae species are present in Vietnam according to PHAM-HOÀNG HÔ (2002). recent contributions by DELOBEL (2010a, b) for Asian Desmodieae and CHAN et al. (2011) for world Desmodium have unveiled an unsuspected diversity of Bruchinae associated with this large group of plants. I report here on three Bruchidius species reared from pod samples of Dendrolobium lanceolatum, Desmodium gangeticum and Tadehagi triquetrum that were collected in Vietnam. This brings the number of seed beetles species known to be associated with Desmodieae in Vietnam to sixteen, and to nineteen (or twenty) species for Asia as a whole. Techniques used for sample collection and rearing were similar to those described earlier (DELOBEL & DELOBEL 2003). Leguminous host plants were identified using the Flora of Vietnam (PHAM-HOÀNG HÔ 2002), and botanical names were updated according to ILDIS (2014); I am greatly indebted to Dr. -

Desmodium Cuspidatum (Muhl.) Loudon Large-Bracted Tick-Trefoil
New England Plant Conservation Program Desmodium cuspidatum (Muhl.) Loudon Large-bracted Tick-trefoil Conservation and Research Plan for New England Prepared by: Lynn C. Harper Habitat Protection Specialist Massachusetts Natural Heritage and Endangered Species Program Westborough, Massachusetts For: New England Wild Flower Society 180 Hemenway Road Framingham, MA 01701 508/877-7630 e-mail: [email protected] • website: www.newfs.org Approved, Regional Advisory Council, 2002 SUMMARY Desmodium cuspidatum (Muhl.) Loudon (Fabaceae) is a tall, herbaceous, perennial legume that is regionally rare in New England. Found most often in dry, open, rocky woods over circumneutral to calcareous bedrock, it has been documented from 28 historic and eight current sites in the three states (Vermont, New Hampshire, and Massachusetts) where it is tracked by the Natural Heritage programs. The taxon has not been documented from Maine. In Connecticut and Rhode Island, the species is reported but not tracked by the Heritage programs. Two current sites in Connecticut are known from herbarium specimens. No current sites are known from Rhode Island. Although secure throughout most of its range in eastern and midwestern North America, D. cuspidatum is Endangered in Vermont, considered Historic in New Hampshire, and watch-listed in Massachusetts. It is ranked G5 globally. Very little is understood about the basic biology of this species. From work on congeners, it can be inferred that there are likely to be no problems with pollination, seed set, or germination. As for most legumes, rhizobial bacteria form nitrogen-fixing nodules on the roots of D. cuspidatum. It is unclear whether there have been any changes in the numbers or distribution of rhizobia capable of forming effective mutualisms with D. -

Atlas of the Flora of New England: Fabaceae
Angelo, R. and D.E. Boufford. 2013. Atlas of the flora of New England: Fabaceae. Phytoneuron 2013-2: 1–15 + map pages 1– 21. Published 9 January 2013. ISSN 2153 733X ATLAS OF THE FLORA OF NEW ENGLAND: FABACEAE RAY ANGELO1 and DAVID E. BOUFFORD2 Harvard University Herbaria 22 Divinity Avenue Cambridge, Massachusetts 02138-2020 [email protected] [email protected] ABSTRACT Dot maps are provided to depict the distribution at the county level of the taxa of Magnoliophyta: Fabaceae growing outside of cultivation in the six New England states of the northeastern United States. The maps treat 172 taxa (species, subspecies, varieties, and hybrids, but not forms) based primarily on specimens in the major herbaria of Maine, New Hampshire, Vermont, Massachusetts, Rhode Island, and Connecticut, with most data derived from the holdings of the New England Botanical Club Herbarium (NEBC). Brief synonymy (to account for names used in standard manuals and floras for the area and on herbarium specimens), habitat, chromosome information, and common names are also provided. KEY WORDS: flora, New England, atlas, distribution, Fabaceae This article is the eleventh in a series (Angelo & Boufford 1996, 1998, 2000, 2007, 2010, 2011a, 2011b, 2012a, 2012b, 2012c) that presents the distributions of the vascular flora of New England in the form of dot distribution maps at the county level (Figure 1). Seven more articles are planned. The atlas is posted on the internet at http://neatlas.org, where it will be updated as new information becomes available. This project encompasses all vascular plants (lycophytes, pteridophytes and spermatophytes) at the rank of species, subspecies, and variety growing independent of cultivation in the six New England states. -

New Combinations in the Desmodium Group of Leguminosae Tribe Desmodieae
J. Jpn. Bot. 93(6): 384–388 (2018) New Combinations in the Desmodium Group of Leguminosae Tribe Desmodieae Hiroyoshi Oa,* and Kazuaki Ob aHerbarium TUS, Botanical Garden, Tohoku University, Sendai, 980-0862 JAPAN; bSchool of Pharmacy, Iwate Medical University, Yahaba, Iwate, 028-3694 JAPAN *Corresponding author: [email protected] (Accepted on July 7, 2018) New combinations in the Desmodium group of Leguminosae tribe Desmodieae are proposed for the following four species based on morphological evidence: Grona retrofl exa (L.) H. Ohashi & K. Ohashi, Pleurolobus flexuosus (Wall. ex Benth.) H. Ohashi & K. Ohashi, P. pryonii (DC.) H. Ohashi & K. Ohashi and Tateishia bolsteri (Merr. & Rolfe) H. Ohashi & K. Ohashi. The lectotype of Hedysarum capitatum Burm. f. is designated. Key words: Desmodium, Grona, Pleurolobus, Tateishia. The Desmodium group of the tribe Heteroloma (Ohashi 1973), but was represented Desmodieae of Leguminosae has recently by a distinct clade and separated from the been reorganized based on morphological Desmodium clade on our phylogenetic trees (cf. and palynological evidence with results of figs. 1–3 in Ohashi et al. 2018b). Desmodium molecular phylogenetic analyses and comprises gangeticum is, therefore, recognized as 28 genera at present (Ohashi et al. 2018a, b). representing an independent monotypic genus Grona, Pleurolobus and Tateishia of the group Pleurolobus. The genus Tateishia was created to are treated in this paper. The genus Grona was accommodate Desmodium concinnum DC. The resurrected from Desmodium for inclusion of species was placed in the section Heteroloma the members of Desmodium sections Nicolsonia (Ohashi 1973), but forms an independent clade and Sagotia (Ohashi and Ohashi 2018). -

Fruits and Seeds of Genera in the Subfamily Faboideae (Fabaceae)
Fruits and Seeds of United States Department of Genera in the Subfamily Agriculture Agricultural Faboideae (Fabaceae) Research Service Technical Bulletin Number 1890 Volume I December 2003 United States Department of Agriculture Fruits and Seeds of Agricultural Research Genera in the Subfamily Service Technical Bulletin Faboideae (Fabaceae) Number 1890 Volume I Joseph H. Kirkbride, Jr., Charles R. Gunn, and Anna L. Weitzman Fruits of A, Centrolobium paraense E.L.R. Tulasne. B, Laburnum anagyroides F.K. Medikus. C, Adesmia boronoides J.D. Hooker. D, Hippocrepis comosa, C. Linnaeus. E, Campylotropis macrocarpa (A.A. von Bunge) A. Rehder. F, Mucuna urens (C. Linnaeus) F.K. Medikus. G, Phaseolus polystachios (C. Linnaeus) N.L. Britton, E.E. Stern, & F. Poggenburg. H, Medicago orbicularis (C. Linnaeus) B. Bartalini. I, Riedeliella graciliflora H.A.T. Harms. J, Medicago arabica (C. Linnaeus) W. Hudson. Kirkbride is a research botanist, U.S. Department of Agriculture, Agricultural Research Service, Systematic Botany and Mycology Laboratory, BARC West Room 304, Building 011A, Beltsville, MD, 20705-2350 (email = [email protected]). Gunn is a botanist (retired) from Brevard, NC (email = [email protected]). Weitzman is a botanist with the Smithsonian Institution, Department of Botany, Washington, DC. Abstract Kirkbride, Joseph H., Jr., Charles R. Gunn, and Anna L radicle junction, Crotalarieae, cuticle, Cytiseae, Weitzman. 2003. Fruits and seeds of genera in the subfamily Dalbergieae, Daleeae, dehiscence, DELTA, Desmodieae, Faboideae (Fabaceae). U. S. Department of Agriculture, Dipteryxeae, distribution, embryo, embryonic axis, en- Technical Bulletin No. 1890, 1,212 pp. docarp, endosperm, epicarp, epicotyl, Euchresteae, Fabeae, fracture line, follicle, funiculus, Galegeae, Genisteae, Technical identification of fruits and seeds of the economi- gynophore, halo, Hedysareae, hilar groove, hilar groove cally important legume plant family (Fabaceae or lips, hilum, Hypocalypteae, hypocotyl, indehiscent, Leguminosae) is often required of U.S. -
Brya Ebenus (L.) DC
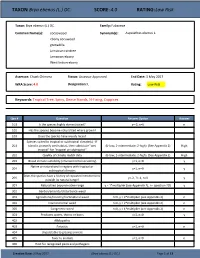
TAXON: Brya ebenus (L.) DC. SCORE: 4.0 RATING: Low Risk Taxon: Brya ebenus (L.) DC. Family: Fabaceae Common Name(s): cocoswood Synonym(s): Aspalathus ebenus L. ebony cocuwood grenadilla Jamaican raintree Jamaican-ebony West Indian-ebony Assessor: Chuck Chimera Status: Assessor Approved End Date: 3 May 2017 WRA Score: 4.0 Designation: L Rating: Low Risk Keywords: Tropical Tree, Spiny, Dense Stands, N-Fixing, Coppices Qsn # Question Answer Option Answer 101 Is the species highly domesticated? y=-3, n=0 n 102 Has the species become naturalized where grown? 103 Does the species have weedy races? Species suited to tropical or subtropical climate(s) - If 201 island is primarily wet habitat, then substitute "wet (0-low; 1-intermediate; 2-high) (See Appendix 2) High tropical" for "tropical or subtropical" 202 Quality of climate match data (0-low; 1-intermediate; 2-high) (See Appendix 2) High 203 Broad climate suitability (environmental versatility) y=1, n=0 y Native or naturalized in regions with tropical or 204 y=1, n=0 y subtropical climates Does the species have a history of repeated introductions 205 y=-2, ?=-1, n=0 y outside its natural range? 301 Naturalized beyond native range y = 1*multiplier (see Appendix 2), n= question 205 y 302 Garden/amenity/disturbance weed 303 Agricultural/forestry/horticultural weed n=0, y = 2*multiplier (see Appendix 2) n 304 Environmental weed n=0, y = 2*multiplier (see Appendix 2) n 305 Congeneric weed n=0, y = 1*multiplier (see Appendix 2) n 401 Produces spines, thorns or burrs y=1, n=0 y 402 Allelopathic 403 Parasitic y=1, n=0 n 404 Unpalatable to grazing animals 405 Toxic to animals y=1, n=0 n 406 Host for recognized pests and pathogens Creation Date: 3 May 2017 (Brya ebenus (L.) DC.) Page 1 of 13 TAXON: Brya ebenus (L.) DC. -

Missouriensis Volume 28 / 29
Missouriensis Volume 28/29 (2008) In this issue: Improved Status of Auriculate False Foxglove (Agalinis auriculata) in Missouri in 2007 Tim E. Smith, Tom Nagel, and Bruce Schuette ......................... 1 Current Status of Yellow False Mallow (Malvastrum hispidum) in Missouri Tim E. Smith.................................................................................... 5 Heliotropium europaeum (Heliotropiaceae) New to Missouri Jay A. Raveill and George Yatskievych ..................................... 10 Melica mutica (Poaceae) New for the Flora of Missouri Alan E. Brant ................................................................................. 18 Schoenoplectus californicus (Cyperaceae) New to Missouri Timothy E. Vogt and Paul M. McKenzie ................................. 22 Flora of Galloway Creek Nature Park, Howell County, Missouri Bill Summers .................................................................................. 27 Journal of the Missouri Native Plant Society Missouriensis, Volume 28/29 2008 1 IMPROVED STATUS OF AURICULATE FALSE FOXGLOVE (AGALINIS AURICULATA) IN MISSOURI IN 2007 Tim E. Smith Missouri Department of Conservation P.O. Box 180, Jefferson City, MO 65102-0180 Tom Nagel Missouri Department of Conservation 701 James McCarthy Drive St. Joseph, MO 64507-2194 Bruce Schuette Missouri Department of Natural Resources Cuivre River State Park 678 State Rt. 147 Troy, MO 63379 Populations of annual plant species are known to have periodic “boom” and “bust” years as well as years when plant numbers more closely approach long-term averages. In tracking populations of plant species of conservation concern (Missouri Natural Heritage Program, 2007), there are sometimes also boom years in the number of reports of new populations. Because of reports of five new populations and a surge in numbers of plants at some previously-known sites, 2007 provided encouraging news for the conservation of the auriculate false foxglove [Agalinis auriculata (Michx.) Blake] in Missouri. -

Download Download
Tropical Natural History 21(1): 41–60, April 2021 ©2021 by Chulalongkorn University Taxonomy of Dendrolobium (Leguminosae) in Thailand WITSANU SAISORN1 AND PRANOM CHANTARANOTHAI2* 1School of Science, Walailak University, Nakhon Si Thammarat 80160, THAILAND 2Department of Biology and Centre of Excellence on Biodiversity (BDC), Faculty of Science, Khon Kaen University, Khon Kaen 40002, THAILAND * Corresponding author. Pranom Chantaranothai ([email protected]) Received: 16 October 2020; Accepted: 21 January 2021 ABSTRACT.– Dendrolobium includes seven species and eight taxa in Thailand, viz. D. baccatum, D. lanceolatum (var. lanceolatum and var. microcarpum), D. olivaceum, D. rugosum, D. thorelii, D. triangulare and D. umbellatum. Four names are lectotypified, including Lespedeza lanceolata, Desmodium wallichii, Desmodium cephalotoides and Desmodium umbellatum var. costatum. Two names are reduced to synonymy, i.e., Lespedeza cambodiana under Dendrolobium lanceolatum and Dendrolobium rugosum var. moniliferum under Dendrolobium rugosum. KEY WORDS: Desmodium, Desmodieae, Fabaceae, Papilionoideae, taxonomy INTRODUCTION Dendrolobium and it is a contribution to the progress of the Flora of Thailand project. Dendrolobium was described by Bentham (1852). The species belonging to MATERIALS AND METHODS this genus have often been treated under Desmodium Desv. in various taxonomic categories, including Desmodium sect. The taxonomic study of genus Eudesmodium (De Candolle, 1825), Dendrolobium in Thailand is based on Desmodium subgen. Dendrolobium (Wight specimens from various herbaria viz. AAU, & Arnott, 1834), and Desmodium sect. ABD, BCU, BK, BKF, BM, BO, C, CMU, Dendrolobium (Bentham, 1864). Nowadays, CMUB, E, FOT, G, G-DC, HN, HNL, Dendrolobium is accepted again in several HNU, K, KEP, KKU, K-W, KYO, L, taxonomic works (e.g., Ohashi, 1973, 1998 NOUL, P, PSU, QBG, SING and TI. -

Don Robinson State Park Species Count: 544
Trip Report for: Don Robinson State Park Species Count: 544 Date: Multiple Visits Jefferson County Agency: MODNR Location: LaBarque Creek Watershed - Vascular Plants Participants: Nels Holmberg, WGNSS, MONPS, Justin Thomas, George Yatskievych This list was compiled by Nels Holmbeg over a period of > 10 years Species Name (Synonym) Common Name Family COFC COFW Acalypha gracilens slender three-seeded mercury Euphorbiaceae 3 5 Acalypha monococca (A. gracilescens var. monococca) one-seeded mercury Euphorbiaceae 3 5 Acalypha rhomboidea rhombic copperleaf Euphorbiaceae 1 3 Acalypha virginica Virginia copperleaf Euphorbiaceae 2 3 Acer rubrum var. undetermined red maple Sapindaceae 5 0 Acer saccharinum silver maple Sapindaceae 2 -3 Achillea millefolium yarrow Asteraceae/Anthemideae 1 3 Actaea pachypoda white baneberry Ranunculaceae 8 5 Adiantum pedatum var. pedatum northern maidenhair fern Pteridaceae Fern/Ally 6 1 Agalinis tenuifolia (Gerardia, A. tenuifolia var. common gerardia Orobanchaceae 4 -3 macrophylla) Ageratina altissima var. altissima (Eupatorium rugosum) white snakeroot Asteraceae/Eupatorieae 2 3 Agrimonia parviflora swamp agrimony Rosaceae 5 -1 Agrimonia pubescens downy agrimony Rosaceae 4 5 Agrimonia rostellata woodland agrimony Rosaceae 4 3 Agrostis perennans upland bent Poaceae/Aveneae 3 1 * Ailanthus altissima tree-of-heaven Simaroubaceae 0 5 * Ajuga reptans carpet bugle Lamiaceae 0 5 Allium canadense var. undetermined wild garlic Liliaceae 2 3 Allium stellatum wild onion Liliaceae 6 5 * Allium vineale field garlic Liliaceae 0 3 Ambrosia artemisiifolia common ragweed Asteraceae/Heliantheae 0 3 Ambrosia bidentata lanceleaf ragweed Asteraceae/Heliantheae 0 4 Amelanchier arborea var. arborea downy serviceberry Rosaceae 6 3 Amorpha canescens lead plant Fabaceae/Faboideae 8 5 Amphicarpaea bracteata hog peanut Fabaceae/Faboideae 4 0 Andropogon gerardii var.