Occurrence of Prey Species Identified from Remains in Regurgitated Pellets Collected from King Shags, 2019 - 2020
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Habitat Use of the Bottlenose Dolphins (Tursiops Truncatus) of Fiordland: Where, Why and the Implications for Management
Habitat use of the bottlenose dolphins (Tursiops truncatus) of Fiordland: Where, why and the implications for management Stephanie M. Bennington A thesis submitted for the degree of Master of Science in Marine Science at the University of Otago, Dunedin, New Zealand. June 2019 Poncho showing off in the beautiful Bowen channel of Dusky Sound photo credit: Steph Bennington i Abstract Understanding the distribution of a species gives important clues about its ecology, and can provide key information and guidance for conservation management. The bottlenose dolphins (Tursiops truncatus) of Fiordland, New Zealand, form three small subpopulations, two of which are mostly resident within separate fjord systems: Doubtful Sound and Dusky Sound. Within these fjords, the dolphins’ distribution and resulting habitat use varies, with high and low use areas, and seasonal variation evident. In this thesis I investigated the distribution patterns of the dolphins, to better understand what drives them and how this relates to the way the dolphins are managed. Specifically, I used Kernel Density Estimation (KDE) from sighting information collected between 2005 and 2018 in Doubtful Sound, and 2009 and 2018 in Dusky Sound, to identify patterns in habitat use over time. Drivers of habitat use were investigated using species distribution models (SDMs), in the form of generalised additive models. Abiotic predictor variables were modelled using long term occurrence data as the response variable. Biotic predictors were included in SDMs for 2018, a year in which I collected data on potential prey and dolphin distribution concurrently. Information on dolphin prey was collected using Baited Underwater Video (BUV). I found that although seasonal variation in habitat use was present, general distribution patterns were consistent through time. -

New Zealand Fishes a Field Guide to Common Species Caught by Bottom, Midwater, and Surface Fishing Cover Photos: Top – Kingfish (Seriola Lalandi), Malcolm Francis
New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing Cover photos: Top – Kingfish (Seriola lalandi), Malcolm Francis. Top left – Snapper (Chrysophrys auratus), Malcolm Francis. Centre – Catch of hoki (Macruronus novaezelandiae), Neil Bagley (NIWA). Bottom left – Jack mackerel (Trachurus sp.), Malcolm Francis. Bottom – Orange roughy (Hoplostethus atlanticus), NIWA. New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing New Zealand Aquatic Environment and Biodiversity Report No: 208 Prepared for Fisheries New Zealand by P. J. McMillan M. P. Francis G. D. James L. J. Paul P. Marriott E. J. Mackay B. A. Wood D. W. Stevens L. H. Griggs S. J. Baird C. D. Roberts‡ A. L. Stewart‡ C. D. Struthers‡ J. E. Robbins NIWA, Private Bag 14901, Wellington 6241 ‡ Museum of New Zealand Te Papa Tongarewa, PO Box 467, Wellington, 6011Wellington ISSN 1176-9440 (print) ISSN 1179-6480 (online) ISBN 978-1-98-859425-5 (print) ISBN 978-1-98-859426-2 (online) 2019 Disclaimer While every effort was made to ensure the information in this publication is accurate, Fisheries New Zealand does not accept any responsibility or liability for error of fact, omission, interpretation or opinion that may be present, nor for the consequences of any decisions based on this information. Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries website at http://www.mpi.govt.nz/news-and-resources/publications/ A higher resolution (larger) PDF of this guide is also available by application to: [email protected] Citation: McMillan, P.J.; Francis, M.P.; James, G.D.; Paul, L.J.; Marriott, P.; Mackay, E.; Wood, B.A.; Stevens, D.W.; Griggs, L.H.; Baird, S.J.; Roberts, C.D.; Stewart, A.L.; Struthers, C.D.; Robbins, J.E. -

Biogenic Habitats on New Zealand's Continental Shelf. Part II
Biogenic habitats on New Zealand’s continental shelf. Part II: National field survey and analysis New Zealand Aquatic Environment and Biodiversity Report No. 202 E.G. Jones M.A. Morrison N. Davey S. Mills A. Pallentin S. George M. Kelly I. Tuck ISSN 1179-6480 (online) ISBN 978-1-77665-966-1 (online) September 2018 Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries websites at: http://www.mpi.govt.nz/news-and-resources/publications http://fs.fish.govt.nz go to Document library/Research reports © Crown Copyright – Fisheries New Zealand TABLE OF CONTENTS EXECUTIVE SUMMARY 1 1. INTRODUCTION 3 1.1 Overview 3 1.2 Objectives 4 2. METHODS 5 2.1 Selection of locations for sampling. 5 2.2 Field survey design and data collection approach 6 2.3 Onboard data collection 7 2.4 Selection of core areas for post-voyage processing. 8 Multibeam data processing 8 DTIS imagery analysis 10 Reference libraries 10 Still image analysis 10 Video analysis 11 Identification of biological samples 11 Sediment analysis 11 Grain-size analysis 11 Total organic matter 12 Calcium carbonate content 12 2.5 Data Analysis of Core Areas 12 Benthic community characterization of core areas 12 Relating benthic community data to environmental variables 13 Fish community analysis from DTIS video counts 14 2.6 Synopsis Section 15 3. RESULTS 17 3.1 -

Trawl Survey of Red Gurnard, John Dory, Tarakihi and Associated Species Off the Bay of Plenty, North Island, February 1999 (KAH9902)
4 IWA Taihoro Nukurangi Trawl survey of red gurnard, John dory, tarakihi and associated species off the Bay of Plenty, North Island, February 1999 (KAH9902) M. Morrison, D. Parkinson Final Research Report for Ministry of Fisheries Research Project INT9803 National Institute of Water and Atmospheric Research November 1999 Final Research Report Report Title: Trawl survey of red gurnard, John dory, terakihi and associated species off the Bay of Plenty, North Island, February 1999 (KAH9902) Authors: M. Morrison, D. Parkinson 1. Date: 15/10/99 2. Contractor: NIWA 3. Project Title: Estimation of inshore fish abundance in the Bay of Plenty using trawl surveys 4. Project Code: INT9803 5. Project Leader: M. Morrison 6. Duration of Project: Start Date 1 October 1998 Expected End Date 30 September 1999 7. Executive Summary: A trawl survey of the Bay of Plenty was successfully completed during February 1999. Seventy eight stations were completed (50 phase 1, 28 phase 2) within 8 depth and area strata. Target c.v.s on biomass estimates were 15% for GUR, 20% for JDO, and 30% for TAR; achieved c.v.s were 14%, 14%, and 27% respectively. Length and weight information was also collected for all other QMS species in the catch, and for leatherjacket, frostfish, kahawai and kingfish. Red gurnard were caught throughout the Bay of Plenty, with relatively large individual catches occurring in the 10-100 m depth range. John dory were less common, with scattered larger catches. Relatively few tarakihi were encountered; most of the tows catching these fish were in deeper water in the middle of the survey area. -

Contrast in the Importance of Arrow Squid As Prey of Male New Zealand Sea Lions and New Zealand Fur Seals at the Snares, Subantarctic New Zealand
Mar Biol (2014) 161:631–643 DOI 10.1007/s00227-013-2366-6 ORIGINAL PAPER Contrast in the importance of arrow squid as prey of male New Zealand sea lions and New Zealand fur seals at The Snares, subantarctic New Zealand Chris Lalas · Trudi Webster Received: 27 April 2013 / Accepted: 3 December 2013 / Published online: 19 December 2013 © Springer-Verlag Berlin Heidelberg 2013 Abstract New Zealand sea lions (Phocarctos hookeri) Introduction are threatened by incidental bycatch in the trawl fishery for southern arrow squid (Nototodarus sloanii). An over- New Zealand sea lions (Phocarctos hookeri), hereafter lap between the fishery and foraging sea lions has previ- referred to as NZ sea lions, have a breeding distribution ously been interpreted as one piece of evidence supporting extending from Otago Peninsula (South Island, New Zea- resource competition for squid. However, there is currently land) south to subantarctic Campbell Island (Fig. 1). The no consensus about the importance of squid in the diet of species is classified as ‘Vulnerable’ by the International New Zealand sea lions. Therefore, we investigated this Union for the Conservation of Nature (IUCN) due to its importance independently of spatial and temporal differ- small population and restricted distribution (IUCN 2012). ences in squid availability through a simultaneous study New Zealand sea lions are the only endemic pinniped spe- with sympatric New Zealand fur seals (Arctocephalus for- cies in New Zealand and 86 % of the entire population steri), a species known to target arrow squid. Diet sampling breed at the subantarctic Auckland Islands (Chilvers et al. at The Snares (48°01′S 166°32′E), subantarctic New Zea- 2007; Chilvers 2008; Fig. -

Determining the Diet of New Zealand King Shag Using DNA Metabarcoding
Determining the diet of New Zealand king shag using DNA metabarcoding New Zealand King Shag, (Leucocarbo carunculatus) on Blumine Island, Marlborough Sounds, New Zealand in 2016 (Wikipedia commons). Aimee van der Reis & Andrew Jeffs Report Prepared For: Department of Conservation, Conservation Services Programme, Project BCBC2019-05. DOC MarineDRAFT Science Advisors Graeme Taylor and Dr Karen Middlemiss. November 2020 Reports from Auckland UniServices Limited should only be used for the purposes for which they were commissioned. If it is proposed to use a report prepared by Auckland UniServices Limited for a different purpose or in a different context from that intended at the time of commissioning the work, then UniServices should be consulted to verify whether the report is being correctly interpreted. In particular it is requested that, where quoted, conclusions given in Auckland UniServices reports should be stated in full. INTRODUCTION The New Zealand king shag (Leucocarbo carunculatus) is an endemic seabird that is classed as nationally endangered (Miskelly et al., 2008). The population is confined to a small number of colonies located around the coastal margins of the outer Marlborough Sounds (South Island, New Zealand); with surveys suggesting the population is currently stable (~800 individuals surveyed in 2020; Aquaculture New Zealand, 2020; Schuckard et al., 2015). Monitoring the colonies has become a priority and research is being conducted to better understand their population dynamics and basic ecology to improve the management of the population, particularly in relation to human activities such as fishing, aquaculture and land use (Fisher & Boren, 2012). The diet of the New Zealand king shag is strongly linked to the waters surrounding their colonies and it has been suggested that anthropogenic activities, such as marine farm structures, may displace foraging habitat that could affect the population of New Zealand king shag (Fisher & Boren, 2012). -

ASFIS ISSCAAP Fish List February 2007 Sorted on Scientific Name
ASFIS ISSCAAP Fish List Sorted on Scientific Name February 2007 Scientific name English Name French name Spanish Name Code Abalistes stellaris (Bloch & Schneider 1801) Starry triggerfish AJS Abbottina rivularis (Basilewsky 1855) Chinese false gudgeon ABB Ablabys binotatus (Peters 1855) Redskinfish ABW Ablennes hians (Valenciennes 1846) Flat needlefish Orphie plate Agujón sable BAF Aborichthys elongatus Hora 1921 ABE Abralia andamanika Goodrich 1898 BLK Abralia veranyi (Rüppell 1844) Verany's enope squid Encornet de Verany Enoploluria de Verany BLJ Abraliopsis pfefferi (Verany 1837) Pfeffer's enope squid Encornet de Pfeffer Enoploluria de Pfeffer BJF Abramis brama (Linnaeus 1758) Freshwater bream Brème d'eau douce Brema común FBM Abramis spp Freshwater breams nei Brèmes d'eau douce nca Bremas nep FBR Abramites eques (Steindachner 1878) ABQ Abudefduf luridus (Cuvier 1830) Canary damsel AUU Abudefduf saxatilis (Linnaeus 1758) Sergeant-major ABU Abyssobrotula galatheae Nielsen 1977 OAG Abyssocottus elochini Taliev 1955 AEZ Abythites lepidogenys (Smith & Radcliffe 1913) AHD Acanella spp Branched bamboo coral KQL Acanthacaris caeca (A. Milne Edwards 1881) Atlantic deep-sea lobster Langoustine arganelle Cigala de fondo NTK Acanthacaris tenuimana Bate 1888 Prickly deep-sea lobster Langoustine spinuleuse Cigala raspa NHI Acanthalburnus microlepis (De Filippi 1861) Blackbrow bleak AHL Acanthaphritis barbata (Okamura & Kishida 1963) NHT Acantharchus pomotis (Baird 1855) Mud sunfish AKP Acanthaxius caespitosa (Squires 1979) Deepwater mud lobster Langouste -

Miscellaneous Demersal Fishes
203 Miscellaneous demersal fishes Capture production by species, fishing areas and countries or areas B-34 Poissons démersaux divers Captures par espèces, zones de pêche et pays ou zones Peces demersales diversos Capturas por especies, áreas de pesca y países o áreas Species, Fishing area Espèce, Zone de pêche 2009 2010 2011 2012 2013 2014 2015 2016 2017 2018 Especie, Área de pesca t t t t t t t t t t Greater argentine Grande argentine ...C Argentina silus 1,23(05)015,03 ARU 27 Germany 30 - - 538 417 1 223 1 066 306 584 1 068 Netherlands ... 2 903 1 562 1 785 1 430 4 267 2 119 2 501 3 438 2 913 Norway 864 664 217 32 258 190 60 146 10 1 27 Fishing area total 894 3 567 1 779 2 355 2 105 5 680 3 245 2 953 4 032 3 982 Species total 894 3 567 1 779 2 355 2 105 5 680 3 245 2 953 4 032 3 982 Argentine Petite argentine Pez plata Argentina sphyraena 1,23(05)015,04 ARY 27 Germany 85 - - 1 143 309 172 1 035 763 790 243 Netherlands 698 248 787 1 662 ... 2 - 1 872 0 Norway 784 671 361 327 996 2 520 3 180 5 830 5 370 7 787 27 Fishing area total 1 567 919 1 148 3 132 1 305 2 694 4 215 6 594 7 032 8 030 Species total 1 567 919 1 148 3 132 1 305 2 694 4 215 6 594 7 032 8 030 Argentines Argentines Argentinas Argentina spp 1,23(05)015,XX ARG 21 Canada 0 2 - - - - - - 2 .. -

Fisheries Resources in Otago Harbour and on the Adjacent Coast
Fisheries resources in Otago Harbour and on the adjacent coast Prepared for Port Otago Limited December 2008 R O Boyd Boyd Fisheries Consultants Ltd 1 Baker Grove Wanaka 9305 NEW ZEALAND Table of Contents Table of Contents i List of Tables and Figures ii Executive Summary iii Fisheries resources in Otago Harbour and on the adjacent coast 1 (Ver 1 – Preliminary) 1 1. Introduction 1 2. Methods 2 2.1. The Fisheries Literature 2 2.2. Fisheries catch and effort data 2 2.3. Consultation with the Fisheries Sector (this section to be revised following further consultation) 2 3. The Fisheries Environments of Otago Harbour and Coastal Otago 3 3.1. Otago Harbour 3 3.2. Coastal Otago 3 4. Fish and Shellfish Fauna of Otago Harbour and Coastal Otago 5 4.1. Otago Harbour 5 4.2. Coastal Otago 6 4.3. Regional and National Significance 7 5. Areas of Importance for Spawning, Egg Laying or Juveniles 8 6. Fisheries Uses of Otago Harbour and Coastal Otago 9 6.1. Recreational Fisheries 9 6.1.1. Otago Harbour 9 6.1.2. Coastal Otago 10 6.2. Commercial Fisheries 10 6.2.1. History and Background to the Commercial Fishery 10 6.2.2. Commercial Fisheries Catch and Effort Data 11 6.2.3. Overview of the Present Otago Commercial Fishery 12 6.2.4. Otago’s Inshore Fisheries 13 Trawl fishery 13 Set net fishery 14 Cod potting 14 Line fishing 14 Paua and kina diving 14 Queen scallops 15 Rock lobster 15 Cockles 15 6.3. Customary Fisheries (this section to be revised following further consultation) 16 7. -
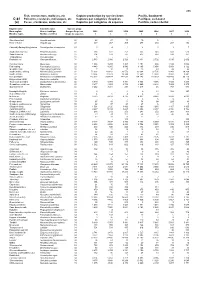
Fish, Crustaceans, Molluscs, Etc Capture Production by Species
495 Fish, crustaceans, molluscs, etc Capture production by species items Pacific, Southwest C-81 Poissons, crustacés, mollusques, etc Captures par catégories d'espèces Pacifique, sud-ouest (a) Peces, crustáceos, moluscos, etc Capturas por categorías de especies Pacífico, sudoccidental English name Scientific name Species group Nom anglais Nom scientifique Groupe d'espèces 2002 2003 2004 2005 2006 2007 2008 Nombre inglés Nombre científico Grupo de especies t t t t t t t Short-finned eel Anguilla australis 22 28 27 13 10 5 ... ... River eels nei Anguilla spp 22 337 267 209 277 210 207 152 Chinook(=Spring=King)salmon Oncorhynchus tshawytscha 23 0 4 1 2 1 1 7 Southern lemon sole Pelotretis flavilatus 31 238 322 251 335 348 608 513 Sand flounders Rhombosolea spp 31 204 193 187 437 514 530 351 Tonguefishes Cynoglossidae 31 3 - - - - - - Flatfishes nei Pleuronectiformes 31 2 580 2 986 2 729 3 431 2 702 3 015 2 602 Common mora Mora moro 32 1 308 1 234 1 403 1 154 986 1 180 1 088 Red codling Pseudophycis bachus 32 4 443 8 265 9 540 8 165 5 854 5 854 6 122 Grenadier cod Tripterophycis gilchristi 32 7 10 13 13 43 29 26 Southern blue whiting Micromesistius australis 32 72 203 43 812 26 576 30 304 32 735 23 943 29 268 Southern hake Merluccius australis 32 13 834 22 623 19 344 12 560 12 858 13 892 8 881 Blue grenadier Macruronus novaezelandiae 32 215 302 209 414 147 032 134 145 119 329 103 489 96 119 Ridge scaled rattail Macrourus carinatus 32 - - - - - 9 14 Thorntooth grenadier Lepidorhynchus denticulatus 32 5 349 5 304 6 341 3 855 4 056 3 725 3 264 Grenadiers, rattails nei Macrouridae 32 3 877 4 253 3 732 2 660 2 848 7 939 8 970 Gadiformes nei Gadiformes 32 3 252 3 281 298 1 217 46 767 886 Broadgilled hagfish Eptatretus cirrhatus 33 2 - 0 0 11 508 347 Sea catfishes nei Ariidae 33 4 6 4 4 4 .. -

Effects of Marine Reserve Protection on Adjacent Non-Protected Populations in New Zealand
Effects of Marine Reserve Protection on Adjacent Non-protected Populations in New Zealand by Daniela Díaz-Guisado A thesis submitted to Victoria University of Wellington in fulfilment of the requirements for the degree of Doctor of Philosophy in Marine Biology. 2014 This thesis was conducted under the supervision of: Associate Professor James J. Bell (Primary supervisor) Victoria University of Wellington, Wellington, New Zealand and Professor Jonathan P.A. Gardner (Secondary supervisor) Victoria University of Wellington, Wellington, New Zealand Abstract Marine reserves (MRs) have been established in many parts of the globe for a variety of reasons and there is an increasing body of evidence that indicates they provide a wide range of benefits that can extend beyond their boundaries. In the present study, the biological effects of protection provided by MRs in New Zealand were evaluated, particularly focusing on the potential impacts of reserves on non-protected areas in terms of export of biomass. First, the biological response of two exploited species to MR protection in New Zealand was quantified by comparing meta-analysis results based on response ratio (RR) analysis and Hedges’ g statistics. Then, effect of MR size and age on those biological responses was determined. Most MRs supported a greater density of larger individuals than unprotected areas. Results indicated that the benefits provided by MRs scale with reserve size. Also, MR age explained a significant amount of the variation in the density and length of both species. Comparison of the performance of RRs with Hedges’ g revealed that RR analysis is an appropriate alternative to Hedges’ g statistic for meta-analyses of MR effectiveness because of its ease of use and interpretation. -

Determining the Diet of New Zealand King Shag Using DNA Metabarcoding
Determining the diet of New Zealand king shag using DNA metabarcoding New Zealand King Shag, (Leucocarbo carunculatus) on Blumine Island, Marlborough Sounds, New Zealand in 2016 (Wikipedia commons). Aimee van der Reis & Andrew Jeffs Report Prepared For: Department of Conservation, Conservation Services Programme, Project BCBC2019-05. DOC Marine Science Advisors Graeme Taylor and Dr Karen Middlemiss. November 2020 Reports from Auckland UniServices Limited should only be used for the purposes for which they were commissioned. If it is proposed to use a report prepared by Auckland UniServices Limited for a different purpose or in a different context from that intended at the time of commissioning the work, then UniServices should be consulted to verify whether the report is being correctly interpreted. In particular it is requested that, where quoted, conclusions given in Auckland UniServices reports should be stated in full. Contents EXECUTIVE SUMMARY ................................................................................................................. 2 1. INTRODUCTION ......................................................................................................................... 3 2. MATERIALS AND METHODS ..................................................................................................... 5 2.1 King shag collection and DNA amplification ........................................................................... 5 2.2 DNA metabarcoding ..............................................................................................................