UCLA Electronic Theses and Dissertations
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nsaids in Cats
ISFM and AAFP Consensus Guidelines Long-term use of NSAIDs in cats Clinical Practice A preprint from the Journal of Feline Medicine and Surgery Volume 12, July 2010 Journal of Feline Medicine and Surgery (2010) 12, 519 doi:10.1016/j.jfms.2010.05.003 EDITORIAL NSAIDs and cats – it’s been a long journey Although the first This issue of JFMS contains the first ever panel have covered much valuable ground: international consensus guidelines ✜ To set the scene they consider how use of NSAIDs on the long-term use of non-steroidal common chronic pain can be in cats, typically was probably anti-inflammatory drugs (NSAIDs) in cats. related to degenerative joint disease, This timely publication, which appears on idiopathic cystitis, trauma and cancer. by Hippocrates, pages 521–538 (doi:10.1016/j.jfms.2010.05.004), ✜ They then explain how and why NSAIDs is a collaborative enterprise by the International can have such positive and, potentially, it has taken until Society of Feline Medicine (ISFM) and negative actions. now for cats to American Association of Feline Practitioners ✜ They consider the best ways of enhancing (AAFP). It has been compiled by a panel of owner and cat compliance, make suggestions gain the benefit of world leaders in the understanding of pain about sensible dosing frequencies, timing of in cats and, without doubt, is essential reading medication and accuracy of dosing, and the long-term use for all small animal veterinary surgeons. emphasize the importance of always using of these drugs. It is interesting to reflect that although the ‘lowest effective dose’. -

List of Withdrawn Drugs
List of withdrawn drugs WRITEN BY LAIMA JONUSIENE To prove that the drug companies make mistakes with our lives we publish this list. Drugs are rushed onto the market for profit. The testing of the drugs is on the major indication. Side effects are NOT tested pre and post. Side effects are observed NOT tested. The expense of pre and post side effect testing is astounding. So it is not done. Side effects are then seen in the public use and a drug is removed from the market after killing or hurting people. There is a special law that prohibits you from suing a drug company for damages unless you can prove they knew it was harmful and sold it anyway. This means there is even less need to test side effects and or report them during the testing process. https://www.youtube.com/watch?v=h7Sd0uBc6eE - How a drug pill is brought to the market. Some drugs have been withdrawn from the market because of risks to the patients. Usually this has been prompted by unexpected adverse effects that were not detected during Phase III clinical trials and were only apparent from postmarketing surveillance data from the wider patient community. This list is not limited to drugs that were ever approved by the FDA. Some of them (Lumiracoxib, Rimonabant, Tolrestat, Ximelagatran and Zimelidine, for example) were approved to be marketed in Europe but had not yet been approved for marketing in the U.S., when side effects became clear and their developers pulled them from the market. Likewise LSD was never approved for marketing in the U.S. -

(CD-P-PH/PHO) Report Classification/Justifica
COMMITTEE OF EXPERTS ON THE CLASSIFICATION OF MEDICINES AS REGARDS THEIR SUPPLY (CD-P-PH/PHO) Report classification/justification of - Medicines belonging to the ATC group M01 (Antiinflammatory and antirheumatic products) Table of Contents Page INTRODUCTION 6 DISCLAIMER 8 GLOSSARY OF TERMS USED IN THIS DOCUMENT 9 ACTIVE SUBSTANCES Phenylbutazone (ATC: M01AA01) 11 Mofebutazone (ATC: M01AA02) 17 Oxyphenbutazone (ATC: M01AA03) 18 Clofezone (ATC: M01AA05) 19 Kebuzone (ATC: M01AA06) 20 Indometacin (ATC: M01AB01) 21 Sulindac (ATC: M01AB02) 25 Tolmetin (ATC: M01AB03) 30 Zomepirac (ATC: M01AB04) 33 Diclofenac (ATC: M01AB05) 34 Alclofenac (ATC: M01AB06) 39 Bumadizone (ATC: M01AB07) 40 Etodolac (ATC: M01AB08) 41 Lonazolac (ATC: M01AB09) 45 Fentiazac (ATC: M01AB10) 46 Acemetacin (ATC: M01AB11) 48 Difenpiramide (ATC: M01AB12) 53 Oxametacin (ATC: M01AB13) 54 Proglumetacin (ATC: M01AB14) 55 Ketorolac (ATC: M01AB15) 57 Aceclofenac (ATC: M01AB16) 63 Bufexamac (ATC: M01AB17) 67 2 Indometacin, Combinations (ATC: M01AB51) 68 Diclofenac, Combinations (ATC: M01AB55) 69 Piroxicam (ATC: M01AC01) 73 Tenoxicam (ATC: M01AC02) 77 Droxicam (ATC: M01AC04) 82 Lornoxicam (ATC: M01AC05) 83 Meloxicam (ATC: M01AC06) 87 Meloxicam, Combinations (ATC: M01AC56) 91 Ibuprofen (ATC: M01AE01) 92 Naproxen (ATC: M01AE02) 98 Ketoprofen (ATC: M01AE03) 104 Fenoprofen (ATC: M01AE04) 109 Fenbufen (ATC: M01AE05) 112 Benoxaprofen (ATC: M01AE06) 113 Suprofen (ATC: M01AE07) 114 Pirprofen (ATC: M01AE08) 115 Flurbiprofen (ATC: M01AE09) 116 Indoprofen (ATC: M01AE10) 120 Tiaprofenic Acid (ATC: -
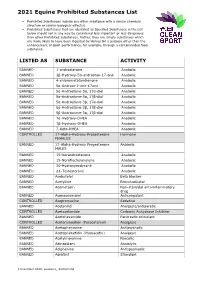
2021 Equine Prohibited Substances List
2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). Prohibited Substances that are identified as Specified Substances in the List below should not in any way be considered less important or less dangerous than other Prohibited Substances. Rather, they are simply substances which are more likely to have been ingested by Horses for a purpose other than the enhancement of sport performance, for example, through a contaminated food substance. LISTED AS SUBSTANCE ACTIVITY BANNED 1-androsterone Anabolic BANNED 3β-Hydroxy-5α-androstan-17-one Anabolic BANNED 4-chlorometatandienone Anabolic BANNED 5α-Androst-2-ene-17one Anabolic BANNED 5α-Androstane-3α, 17α-diol Anabolic BANNED 5α-Androstane-3α, 17β-diol Anabolic BANNED 5α-Androstane-3β, 17α-diol Anabolic BANNED 5α-Androstane-3β, 17β-diol Anabolic BANNED 5β-Androstane-3α, 17β-diol Anabolic BANNED 7α-Hydroxy-DHEA Anabolic BANNED 7β-Hydroxy-DHEA Anabolic BANNED 7-Keto-DHEA Anabolic CONTROLLED 17-Alpha-Hydroxy Progesterone Hormone FEMALES BANNED 17-Alpha-Hydroxy Progesterone Anabolic MALES BANNED 19-Norandrosterone Anabolic BANNED 19-Noretiocholanolone Anabolic BANNED 20-Hydroxyecdysone Anabolic BANNED Δ1-Testosterone Anabolic BANNED Acebutolol Beta blocker BANNED Acefylline Bronchodilator BANNED Acemetacin Non-steroidal anti-inflammatory drug BANNED Acenocoumarol Anticoagulant CONTROLLED Acepromazine Sedative BANNED Acetanilid Analgesic/antipyretic CONTROLLED Acetazolamide Carbonic Anhydrase Inhibitor BANNED Acetohexamide Pancreatic stimulant CONTROLLED Acetominophen (Paracetamol) Analgesic BANNED Acetophenazine Antipsychotic BANNED Acetophenetidin (Phenacetin) Analgesic BANNED Acetylmorphine Narcotic BANNED Adinazolam Anxiolytic BANNED Adiphenine Antispasmodic BANNED Adrafinil Stimulant 1 December 2020, Lausanne, Switzerland 2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). -

The Use of a Subanesthetic Infusion of Intravenous Ketamine to Allow Withdrawal of Medically Prescribed Opioids in People with Chronic Pain, Opioid Tolerance And
bs_bs_banner Pain Medicine 2012; 13: 1524–1525 Wiley Periodicals, Inc. The Use of a Subanesthetic Infusion of Intravenous Ketamine to Allow Withdrawal of Medically Prescribed Opioids in People with Chronic Pain, Opioid Tolerance and Hyperalgesia: Outcome at 6 Monthspme_1486 1524..1525 To the Editor, prior to detoxification. None of the patients had dorsal column stimulators or pumps. The problems of opioid tolerance [1], hyperalgesia (and other side effects of prescribed opioids), and the difficul- The questionnaire involved patients scoring five questions ties involved with opioid withdrawal, are rapidly becoming on an analogue scale from 0 (disagree) through to 5 an enormous health problem. Ketamine, in addition to the (strongly agree). Patients were asked whether they had potential to reduce pain for some weeks or months any further comments regarding the treatment, and to give postinfusion [2], has been shown to ameliorate symptoms a list of current medications (Table 1). of opioid withdrawal [3]. The responses show that: Patients with chronic pain taking medically prescribed opioids who had developed opioid-related tolerance and • While having the infusion was not pleasant, it was tol- probable hyperalgesia were admitted for 5 days of sub- erated by most. anesthetic infusion of ketamine to assist in complete with- • The patients felt better initially. drawal of opioids.1 The aim was to withdraw all opioid • Eight of 11 continued to feel better at 2 months. medication for a period of at least 2 weeks to allow toler- • At 6 months, three of the patients continued to feel well ance to minimize and review progress over 6 months. -
FDA Briefing Document
1 FDA Briefing Document Psychopharmacologic Drugs Advisory Committee (PDAC) and Drug Safety and Risk Management (DSaRM) Advisory Committee Meeting February 12, 2019 Agenda Topic: The committees will discuss the efficacy, safety, and risk-benefit profile of New Drug Application (NDA) 211243, esketamine 28 mg single-use nasal spray device, submitted by Janssen Pharmaceuticals, Inc., for the treatment of treatment-resistant depression 2 The attached package contains background information prepared by the Food and Drug Administration (FDA) for the panel members of the advisory committee. The FDA background package often contains assessments and/or conclusions and recommendations written by individual FDA reviewers. Such conclusions and recommendations do not necessarily represent the final position of the individual reviewers, nor do they necessarily represent the final position of the Review Division or Office. We have brought New Drug Application 211243, esketamine for the treatment of treatment-resistant depression to this Advisory Committee to gain the Committee’s insights and opinions, and the background package may not include all issues relevant to the final regulatory recommendation and instead is intended to focus on issues identified by the Agency for discussion by the advisory committee. The FDA will not issue a final determination on the issues at hand until input from the advisory committee process has been considered and all reviews have been finalized. The final determination may be affected by issues not discussed at the advisory -

The NCGC Pharmaceutical Collection
PERSPECTIVE PHARMACOLOGY indications (such as in the case of pregaba- lin, an antiepileptic drug that was also found The NCGC Pharmaceutical Collection: to be useful for treating neuropathic pain) (6–8) or the withdrawal of marketing autho- A Comprehensive Resource of Clinically rization (which occurred for fen uramine, originally used to suppress appetite but Approved Drugs Enabling Repurposing and then found to lead to cardiotoxicity) (9, 10). Extension of the clinical use of a drug to a Chemical Genomics new indication has historically occurred via serendipitous clinical observation (for ex- Ruili Huang,* Noel Southall,* Yuhong Wang, Adam Yasgar, Paul Shinn, ample, the observation that sildena l, which Ajit Jadhav, Dac-Trung Nguyen, Christopher P. Austin was developed for treating hypertension, was also useful for treating erectile dysfunc- Small-molecule compounds approved for use as drugs may be “repurposed” for new tion) but more recently has occurred via a indications and studied to determine the mechanisms of their bene cial and adverse logical connection of a disease’s pathophysi- e ects. A comprehensive collection of all small-molecule drugs approved for human ology to a drug’s target. Examples of this sce- use would be invaluable for systematic repurposing across human diseases, particularly nario include (i) losartan, a drug developed for rare and neglected diseases, for which the cost and time required for development of a new chemical entity are often prohibitive. Previous e orts to build such a compre- to treat hypertension that might also be use- hensive collection have been limited by the complexities, redundancies, and semantic ful for treating Marfan syndrome because of inconsistencies of drug naming within and among regulatory agencies worldwide; a lack its e ects on transforming growth factor–β, of clear conceptualization of what constitutes a drug; and a lack of access to physical which plays a role in this condition, and (ii) samples. -

Summary of Product Characteristics
Health Products Regulatory Authority Summary of Product Characteristics 1 NAME OF THE VETERINARY MEDICINAL PRODUCT TOLFEDOL, 40 mg/ml, solution for injection for cattle, pigs, cats and dogs 2 QUALITATIVE AND QUANTITATIVE COMPOSITION Each ml contains: Active substance: Tolfenamic acid ……………………………………………… 40 mg Excipients: Benzyl alcohol (E1519)… …… … …… … …… … …… … …… ..10.4 mg Sodium formaldehyde sulphoxylate………………………….. 5 mg For the full list of excipients, see section 6.1. 3 PHARMACEUTICAL FORM Solution for injection. A clear yellowish solution. 4 CLINICAL PARTICULARS 4.1 Target Species Cattle, pigs, cats and dogs. 4.2 Indications for use, specifying the target species In cattle, as an adjunct in the treatment of pneumonia by improving general conditions and nasal discharge and as an adjunct in the treatment of acute mastitis. In pigs, as an adjunct in the treatment of Metritis Mastitis Agalactia syndrome. In dogs : for the treatment of inflammation associated with musculo-skeletal disorders and for the reduction of post-operative pain. In cats : as an adjunct in the treatment of upper respiratory disease in association with antimicrobial therapy, if appropriate. 4.3 Contraindications Do not use in cases of cardiac disease. Do not use in cases of impaired hepatic function or acute renal insufficiency. Do not use in cases of ulceration or digestive bleeding, in case of blood dyscrasia. Do not inject intramuscularly in cats. Do not use in known cases of hypersensitivity to the active substance or to any of the excipients. Do not use in dehydrated, hypovolaemic or hypotonic animals (due to its potential risk of increasing renal toxicity). Do not administer other steroidal or non – steroidal anti – inflammatory drugs concurrently or within 24 hours of each other. -

Genome Sequence Variability Predicts Drug Precautions and Withdrawals from the Market
RESEARCH ARTICLE Genome Sequence Variability Predicts Drug Precautions and Withdrawals from the Market Kye Hwa Lee1, Su Youn Baik1, Soo Youn Lee1, Chan Hee Park1, Paul J. Park3, Ju Han Kim1,2* 1 Seoul National University Biomedical Informatics (SNUBI), Division of Biomedical Informatics, Seoul National University College of Medicine, Seoul 110799, Korea, 2 Biomedical Informatics Training and Education Center (BITEC), Seoul National University Hospital, Seoul 110744, Korea, 3 Department of a11111 Physiology and Cell Biology, University of Nevada School of Medicine, Reno, Nevada, United States of America * [email protected] Abstract OPEN ACCESS Despite substantial premarket efforts, a significant portion of approved drugs has been Citation: Lee KH, Baik SY, Lee SY, Park CH, Park PJ, Kim JH (2016) Genome Sequence Variability withdrawn from the market for safety reasons. The deleterious impact of nonsynonymous Predicts Drug Precautions and Withdrawals from substitutions predicted by the SIFT algorithm on structure and function of drug-related pro- the Market. PLoS ONE 11(9): e0162135. teins was evaluated for 2504 personal genomes. Both withdrawn (n = 154) and precaution- doi:10.1371/journal.pone.0162135 ary (Beers criteria (n = 90), and US FDA pharmacogenomic biomarkers (n = 96)) drugs Editor: Jinn-Moon Yang, National Chiao Tung showed significantly lower genomic deleteriousness scores (P < 0.001) compared to others University College of Biological Science and Technology, TAIWAN (n = 752). Furthermore, the rates of drug withdrawals and precautions correlated signifi- cantly with the deleteriousness scores of the drugs (P < 0.01); this trend was confirmed for Received: April 7, 2016 all drugs included in the withdrawal and precaution lists by the United Nations, European Accepted: July 22, 2016 Medicines Agency, DrugBank, Beers criteria, and US FDA. -

A Machine Learning Approach Predicts Tissue-Specific Drug Adverse 2 Events 3 4 Neel S
bioRxiv preprint doi: https://doi.org/10.1101/288332; this version posted March 24, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 A Machine Learning Approach Predicts Tissue-Specific Drug Adverse 2 Events 3 4 Neel S. Madhukar*1,2,3,4, Kaitlyn Gayvert*1,2,3,4, Coryandar Gilvary1,2,3,4, Olivier Elemento1,2,3,4 5 6 1 HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine, Dept. of 7 Physiology and Biophysics, Weill Cornell Medicine, New York, NY 10065, USA; 8 2 Caryl and Israel Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 9 10065, USA; 10 3 Sandra and Edward Meyer Cancer Center, Weill Cornell Medicine, New York, NY 10065, USA; 11 4 Tri-Institutional Training Program in Computational Biology and Medicine, New York, NY 10065, USA; 12 13 * co-first authors 14 15 Correspondence: Olivier Elemento ([email protected]) 16 17 ABSTRACT 18 19 One of the main causes for failure in the drug development pipeline or withdrawal post 20 approval is the unexpected occurrence of severe drug adverse events. Even though such 21 events should be detected by in vitro, in vivo, and human trials, they continue to 22 unexpectedly arise at different stages of drug development causing costly clinical trial 23 failures and market withdrawal. Inspired by the “moneyball” approach used in baseball to 24 integrate diverse features to predict player success, we hypothesized that a similar 25 approach could leverage existing adverse event and tissue-specific toxicity data to learn 26 how to predict adverse events. -

Correlation Between Drug Market Withdrawals and Socioeconomic, Health, and Welfare Indicators Worldwide
ORIGINAL ARTICLE Medicine General & Social Medicine http://dx.doi.org/10.3346/jkms.2015.30.11.1567 • J Korean Med Sci 2015; 30: 1567-1576 Correlation between Drug Market Withdrawals and Socioeconomic, Health, and Welfare Indicators Worldwide Kye Hwa Lee, Grace Juyun Kim, The relationship between the number of withdrawn/restricted drugs and socioeconomic, and Ju Han Kim health, and welfare indicators were investigated in a comprehensive review of drug regulation information in the United Nations (UN) countries. A total of of 362 drugs were Seoul National University Biomedical Informatics withdrawn and 248 were restricted during 1950-2010, corresponding to rates of 12.02 ± (SNUBI) and Systems Biomedical Informatics ± ± Research Center, Division of Biomedical Informatics, 13.07 and 5.77 8.69 (mean SD), respectively, among 94 UN countries. A Seoul National University College of Medicine, socioeconomic, health, and welfare analysis was performed for 33 OECD countries for Seoul, Korea which data were available regarding withdrawn/restricted drugs. The gross domestic product (GDP) per capita, GDP per hour worked, health expenditure per GDP, and elderly Received: 23 April 2015 Accepted: 10 July 2015 population rate were positively correlated with the numbers of withdrawn and restricted drugs (P < 0.05), while the out-of-pocket health expenditure payment rate was negatively Address for Correspondence: correlated. The number of restricted drugs was also correlated with the rate of drug-related Ju Han Kim, MD deaths (P < 0.05). The World Bank data cross-validated the findings of 33 OECD countries. Division of Biomedical Informatics, Systems Biomedical Informatics Research Center, Seoul National University College The lists of withdrawn/restricted drugs showed markedly poor international agreement of Medicine, 103 Daehak-ro, Jongno-gu, Seoul 03080, Korea between them (Fleiss’s kappa = -0.114). -

Cheminformatics Approaches to Drug Discovery: from Knowledgebases to Toxicity Prediction and Promiscuity Assessment
Cheminformatics Approaches to Drug Discovery: From Knowledgebases to Toxicity Prediction and Promiscuity Assessment Inaugural-Dissertation to obtain the academic degree Doctor rerum naturalium (Dr. rer. nat.) submitted to the Department of Biology, Chemistry and Pharmacy of Freie Universität Berlin by VISHAL BABU SIRAMSHETTY from Hyderabad, India 2018 This research work was conducted from December 2014 to June 2018 under the supervision of PD Dr. Robert Preissner at the Charité – Universitätsmedizin Berlin. 1. Reviewer: PD Dr. Robert Preissner (Charité – Universitätsmedizin Berlin) 2. Reviewer: Prof. Dr. Gerhard Wolber (Freie Universität Berlin) Date of defense: 09.01.2019 Acknowledgements I would like to take this opportunity to thank all those people who have accompanied me during the last years and contributed in many ways to the completion of this dissertation. Firstly, I would like to thank my supervisor PD Dr. Robert Preissner for his guidance, encouragement, and support throughout my doctoral study. The last four years had a significant impact on my life. I believe I evolved as a researcher as well as a social human being. I feel privileged to participate in international conferences, meet some pioneers in the field and present my work. Dear Robert, I sincerely thank you for providing the freedom to explore my interests and the trust you placed on me. I would also like to thank Prof. Dr. Gerhard Wolber for being the co-referent of my thesis. I extend my gratitude to all my colleagues of the Structural Bioinformatics Group at Charité for being so kind and for providing a friendly and interactive working atmosphere. Many thanks to Malgorzata Drwal, Priyanka Banerjee, Andreas Oliver Eckert, Björn Oliver Gohlke, and Janette Nickel-Seeber for the highly productive and pleasant collaborations within the group.