A Molecular Systematic Study of the African Endemic Cycads
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
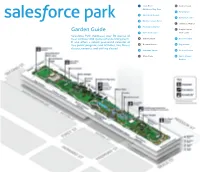
Salesforce Park Garden Guide
Start Here! D Central Lawn Children’s Play Area Garden Guide6 Palm Garden 1 Australian Garden Start Here! D Central Lawn Salesforce Park showcases7 California over Garden 50 species of Children’s Play Area 2 Mediterraneantrees and Basin over 230 species of understory plants. 6 Palm Garden -ã ¼ÜÊ ÊăØÜ ØÊèÜãE úØƀØÊèÃJapanese Maples ¼ÃØ Ê¢ 1 Australian Garden 3 Prehistoric¢ØÕ輫ÕØÊ£ØÂÜÃã«ó«ã«Üŧ¼«¹ĆãÃÜÜ Garden 7 California Garden ¼ÜÜÜŧÊÃØãÜŧÃØ¢ã«Ã£¼ÜÜÜũF Amphitheater Garden Guide 2 Mediterranean Basin 4 Wetland Garden Main Lawn E Japanese Maples Salesforce Park showcases over 50 species of 3 Prehistoric Garden trees and over 230 species of understory plants. A Oak Meadow 8 Desert Garden F Amphitheater It also offers a robust year-round calendar of 4 Wetland Garden Main Lawn free public programs and activities, like fitness B Bamboo Grove 9 Fog Garden Desert Garden classes, concerts, and crafting classes! A Oak Meadow 8 5 Redwood Forest 10 Chilean Garden B Bamboo Grove 9 Fog Garden C Main Plaza 11 South African 10 Chilean Garden Garden 5 Redwood Forest C Main Plaza 11 South African Garden 1 Children’s Australian Play Area Garden ABOUT THE GARDENS The botanist aboard the Endeavor, Sir Joseph Banks, is credited with introducing many plants from Australia to the western world, and many This 5.4 acre park has a layered soil system that plants today bear his name. balances seismic shifting, collects and filters storm- water, and irrigates the gardens. Additionally, the soil Native to eastern Australia, Grass Trees may grow build-up and dense planting help offset the urban only 3 feet in 100 years, and mature plants can be heat island effect by lowering the air temperature. -

Cycad Species List
Mailing Address: E mail: [email protected] 3233 Brant St. Phone: (619) 291-4605 San Diego CA, 92103-5503 Jungle Music Fax: (619) 574-1595 Nursery Location: 450 Ocean View Ave. Palms & Cycads We’re on the Web! Encinitas CA www.junglemusic.net The palm and cycad source since 1977 Cycad Availability SPRING/SUMMER 2006 Directions Leucadia Blvd Take Freeway 5 to Encinitas (10 minutes south of Oceanside, 30 minutes north of San Diego). N Ocean View Ave Ave View Ocean e Exit Leucadia Blvd West (toward ocean). v A W E s Immediate left on Orpheus Ave, u e h left on Union St, p S r Right on Ocean View Ave O Union St Location: 450 Ocean View Ave, Encinitas, CA (next to the “Monarch Program”) The following is a listing of species available as of APRIL 2006. Call for sizes and prices available. Many species are extremely limited in availability. Some species may have sold out by the time you get this listing. Some cycad species are given as a “locality”. For visits to the nursery, always call first. Mail orders are possible to most areas within the United States with minimum order of $200 plus s/h. Thank you for your interest. Cycads are for U.S. domestic use only and cannot be shipped internationally. Phil Bergman, Owner The purpose of this listing is to give palm and cycad enthusiasts a list of the species offered at this time. Sizes and prices are not included herein as availability changes frequently and creating such a listing would be far too cumber- some. -

Somatic Embryogenesis and Regeneration of Endangered Cycad Species
Somatic Embryogenesis and Regeneration of Endangered Cycad Species R.E. Litz and P.A. Moon V.M. Chavez Avila Tropical Research and Education Center Jardin Botanico, Instituto de Biologia University of Florida Universidad Nacional Autonoma de Mexico 18905 SW 280 Street Apartado Postal 70-614 Homestead FL, 33031-3314 04510 Mexico DF USA Mexico Keywords: Somatic embryo, gymnosperm, Cycadales, conservation Abstract The Cycadales (Gymnospermae) include some of the world's most endangered and rare plant species. Many of the cycad species are known only as single specimen trees (e.g., Encephalartos woodii), as very small populations in the wild (e.g., Ceratozamia hildae) or have become extinct in the wild (e.g., Ceratozamia euryphyllidia). All cycads are dioecious, so that seed production is no longer possible with the rarest of the species. Conditions for induction of embryogenic cultures from leaves of mature phase trees of several species in the family Zamiaceae have been reported, and plants have been regenerated from somatic embryos. Embryogenic cultures of two species have been successfully cryopreserved. These strategies should contribute to the conservation of these endangered species and could lay the basis for commercial propagation of these beautiful but rare plants. INTRODUCTION The Cycadales represent the most ancient surviving group of higher plants, having arisen during the Permian era and flourished in the Mesozoic and Jurassic periods. They have been referred to as "living fossils" (Gilbert, 1984). Norstog (1987) considered that the cycads are unique for the study of the evolution of development in higher plants. There are only three extant cycad families, the Cycadaceae, Stangeriaceae and Zamiaceae, and these contain approximately 224 species. -

Chemical Element Concentrations of Cycad Leaves: Do We Know Enough?
horticulturae Review Chemical Element Concentrations of Cycad Leaves: Do We Know Enough? Benjamin E. Deloso 1 , Murukesan V. Krishnapillai 2 , Ulysses F. Ferreras 3, Anders J. Lindström 4, Michael Calonje 5 and Thomas E. Marler 6,* 1 College of Natural and Applied Sciences, University of Guam, Mangilao, GU 96923, USA; [email protected] 2 Cooperative Research and Extension, Yap Campus, College of Micronesia-FSM, Colonia, Yap 96943, Micronesia; [email protected] 3 Philippine Native Plants Conservation Society Inc., Ninoy Aquino Parks and Wildlife Center, Quezon City 1101, Philippines; [email protected] 4 Plant Collections Department, Nong Nooch Tropical Botanical Garden, 34/1 Sukhumvit Highway, Najomtien, Sattahip, Chonburi 20250, Thailand; [email protected] 5 Montgomery Botanical Center, 11901 Old Cutler Road, Coral Gables, FL 33156, USA; [email protected] 6 Western Pacific Tropical Research Center, University of Guam, Mangilao, GU 96923, USA * Correspondence: [email protected] Received: 13 October 2020; Accepted: 16 November 2020; Published: 19 November 2020 Abstract: The literature containing which chemical elements are found in cycad leaves was reviewed to determine the range in values of concentrations reported for essential and beneficial elements. We found 46 of the 358 described cycad species had at least one element reported to date. The only genus that was missing from the data was Microcycas. Many of the species reports contained concentrations of one to several macronutrients and no other elements. The cycad leaves contained greater nitrogen and phosphorus concentrations than the reported means for plants throughout the world. Magnesium was identified as the macronutrient that has been least studied. -

Exposing the Illegal Trade in Cycad Species (Cycadophyta: Encephalartos) at Two Traditional Medicine Markets in South Africa Using DNA Barcoding1 J
771 ARTICLE Exposing the illegal trade in cycad species (Cycadophyta: Encephalartos) at two traditional medicine markets in South Africa using DNA barcoding1 J. Williamson, O. Maurin, S.N.S. Shiba, H. van der Bank, M. Pfab, M. Pilusa, R.M. Kabongo, and M. van der Bank Abstract: Species in the cycad genus Encephalartos are listed in CITES Appendix I and as Threatened or Protected Species in terms of South Africa’s National Environmental Management: Biodiversity Act (NEM:BA) of 2004. Despite regulations, illegal plant harvesting for medicinal trade has continued in South Africa and resulted in declines in cycad populations and even complete loss of sub-populations. Encephalartos is traded at traditional medicine markets in South Africa in the form of bark strips and stem sections; thus, determining the species traded presents a major challenge due to a lack of characteristic plant parts. Here, a case study is presented on the use of DNA barcoding to identify cycads sold at the Faraday and Warwick traditional medicine markets in Johannesburg and Durban, respectively. Market samples were sequenced for the core DNA barcodes (rbcLa and matK) as well as two additional regions: nrITS and trnH-psbA. The barcoding database for cycads at the University of Johannesburg was utilized to assign query samples to known species. Three approaches were followed: tree-based, similarity-based, and character-based (BRONX) methods. Market sam- ples identified were Encephalartos ferox (Near Threatened), Encephalartos lebomboensis (Endangered), Encephalartos natalensis (Near Threatened), Encephalartos senticosus (Vulnerable), and Encephalartos villosus (Least Concern). Results from this study are crucial for making appropriate assessments and decisions on how to manage these markets. -

Encephalartos Aemulans (Zamiaceae), a New Species From
S. Afr.J. Bot., 1990, 56(2): 239-243 239 EncephaJartos aemuJans (Zamiaceae), a new species from northern Natal P. Vorster Botany Department, University of Stellenbosch, Stellenbosch, 7600 Republic of South Africa Accepted 12 December 1989 Encephalartos aemulans Vorster is described from the Ngotshe district in northern Natal. It resembles E. natalensis R.A. Dyer & Verdoorn by the tuberculose bullae of the female cones, and E. lebomboensis Verdoorn by the relatively narrow leaflets which are reduced to prickles towards the base of the frond; but differs from both by the thick indumentum covering the cone surfaces, the sessile male cones without drooping beaks on the bullae, and by the morphological similarity between the submature male and female cones. Encephalartos aemulans word beskryf vanuit die Ngotshe-distrik in Noord-Natal. Dit stem ooreen met E. natalensis R.A. Dyer & Verdoorn op grond van die vratterige bullae van die vroulike keels, en E. lebomboensis Verdoorn op grond van die relatief smal pinnae wat reduseer is tot dorinkies na die basis van die blaar toe; maar dit verskil van beide deur die digte haarbedekking van die keeloppervlaktes, die sittende manlike keels waarvan die bullae nie afbuigende snawels vorm nie, en deur die morfologiese ooreenkoms tussen die byna volwasse manlike en vroulike keels. Keywords: Encephalartos, new species, Zamiaceae A critical study of Encephalartos natalensis R.A. Dyer & both ends, apices oblique and pungent, margins very Verdoorn led to the conclusion that plants from northern slightly revolute (more conspicuously so in dried than Natal represent a distinct and undescribed species: fresh material) with 1-3 teeth on upper margin and 1-2 on lower margin . -

Download the PDF File
ISSN 2473-442X CONTENTS Message from Dr. Patrick Griffith, Co-chair, IUCN/SSC CSG 3 Official newsletter of IUCN/SSC Cycad Specialist Group Botanic Garden: In Focus Vol. IV I Issue 2 I December 2019 Montgomery Botanical Center’s Cycad Collection – Focus on research and conservation 5 Michael Calonje & Patrick Griffith Feature Articles Towards an approach for the conservation and illegal trade prevention of South Africa’s endangered Encephalartos spp. 10 James A. R. Clugston, Michelle Van Der Bankand Ronny M. Kobongo Fire is the most important threat for conservation of Dioon merolae (espadaña) in the hill Nambiyigua, municipality of Villaflores, Chiapas, Mexico 13 Miguel Angel Pérez-Farrera & Mauricio Martínez Martínez Ex-situ Cycad Conservation [1]: Public and Private Collections 16 Chip Jones & JS Khuraijam The Cycad Specialist Group (CSG) is a component of the IUCN Species Research and Conservation News Survival Commission (IUCN/SSC). It consists of a group of volunteer The Cycad Extinction Crisis in South Africa 19 experts addressing conservation Wynand van Eeden & Tim Gregory issues related to cycads, a highly What is Ceratozamia becerrae ? 21 threatened group of land plants. The Andrew P. Vovides, Miguel Angel Pérez-Farrera & José Said Gutiérrez-Ortega CSG exists to bring together the world’s cycad conservation expertise, Preliminary Finding: Seed longevity of Encephalartos in controlled storage 23 and to disseminate this expertise to Ngawethu Ngaka and Phakamani Xaba organizations and agencies which can use this guidance to advance cycad Meeting Reports conservation. 2nd Nong Nooch Cycad Horticulture Workshop 25 Official website of CSG: Anders Lindstrom http://www.cycadgroup.org/ Plant Conservation Genetics Workshop 26 Co-Chairs Caroline Iacuaniello, Stephanie Steele & Christy Powell John Donaldson Patrick Griffith CSG Members 28 Vice Chairs Michael Calonje Cristina Lopez-Gallego Red List Authority Coordinator De Wet Bosenberg CSG Newsletter Committee JS Khuraijam, Editor Irene Terry Andrew P. -

Coevolution of Cycads and Dinosaurs George E
Coevolution of cycads and dinosaurs George E. Mustoe* INTRODUCTION TOXICOLOGY OF EXTANT CYCADS cycads suggests that the biosynthesis of ycads were a major component of Illustrations in textbooks commonly these compounds was a trait that C forests during the Mesozoic Era, the depict herbivorous dinosaurs browsing evolved early in the history of the shade of their fronds falling upon the on cycad fronds, but biochemical evi- Cycadales. Brenner et al. (2002) sug- scaly backs of multitudes of dinosaurs dence from extant cycads suggests that gested that macrozamin possibly serves a that roamed the land. Paleontologists these reconstructions are incorrect. regulatory function during cycad have long postulated that cycad foliage Foliage of modern cycads is highly toxic growth, but a strong case can be made provided an important food source for to vertebrates because of the presence that the most important reason for the reptilian herbivores, but the extinction of two powerful neurotoxins and carcin- evolution of cycad toxins was their of dinosaurs and the contemporaneous ogens, cycasin (methylazoxymethanol- usefulness as a defense against foliage precipitous decline in cycad popula- beta-D-glucoside) and macrozamin (beta- predation at a time when dinosaurs were tions at the close of the Cretaceous N-methylamine-L-alanine). Acute symp- the dominant herbivores. The protective have generally been assumed to have toms triggered by cycad foliage inges- role of these toxins is evidenced by the resulted from different causes. Ecologic tion include vomiting, diarrhea, and seed dispersal characteristics of effects triggered by a cosmic impact are abdominal cramps, followed later by loss modern cycads. a widely-accepted explanation for dino- of coordination and paralysis of the saur extinction; cycads are presumed to limbs. -

Comparative Biology of Cycad Pollen, Seed and Tissue - a Plant Conservation Perspective
Bot. Rev. (2018) 84:295–314 https://doi.org/10.1007/s12229-018-9203-z Comparative Biology of Cycad Pollen, Seed and Tissue - A Plant Conservation Perspective J. Nadarajan1,2 & E. E. Benson 3 & P. Xaba 4 & K. Harding3 & A. Lindstrom5 & J. Donaldson4 & C. E. Seal1 & D. Kamoga6 & E. M. G. Agoo7 & N. Li 8 & E. King9 & H. W. Pritchard1,10 1 Royal Botanic Gardens, Kew, Wakehurst Place, Ardingly, West Sussex RH17 6TN, UK; e-mail: [email protected] 2 The New Zealand Institute for Plant & Food Research Ltd, Private Bag 11600, Palmerston North 4442, New Zealand; e-mail [email protected] 3 Damar Research Scientists, Damar, Cuparmuir, Fife KY15 5RJ, UK; e-mail: [email protected]; [email protected] 4 South African National Biodiversity Institute, Kirstenbosch National Botanical Garden, Cape Town, Republic of South Africa; e-mail: [email protected]; [email protected] 5 Nong Nooch Tropical Botanical Garden, Chonburi 20250, Thailand; e-mail: [email protected] 6 Joint Ethnobotanical Research Advocacy, P.O.Box 27901, Kampala, Uganda; e-mail: [email protected] 7 De La Salle University, Manila, Philippines; e-mail: [email protected] 8 Fairy Lake Botanic Garden, Shenzhen, Guangdong, People’s Republic of China; e-mail: [email protected] 9 UNEP-World Conservation Monitoring Centre, Cambridge, UK; e-mail: [email protected] 10 Author for Correspondence; e-mail: [email protected] Published online: 5 July 2018 # The Author(s) 2018 Abstract Cycads are the most endangered of plant groups based on IUCN Red List assessments; all are in Appendix I or II of CITES, about 40% are within biodiversity ‘hotspots,’ and the call for action to improve their protection is long- standing. -

35 Ideal Landscape Cycads
3535 IdealIdeal LandscapeLandscape CycadsCycads Conserve Cycads by Growing Them -- Preservation Through Propagation Select Your Plant Based on these Features: Exposure: SunSun ShadeShade ☻☻ ColdCold☻☻ Filtered/CoastalFiltered/Coastal SunSun ▲▲ Leaf Length and Spread: Compact, Medium or Large? Growth Rate and Ultimate Plant Size Climate: Subtropical, Mediterranean, Temperate? Dry or Moist? Leaves -- Straight or Arching? Ocean-Loving, Salt-Tolerant, Wind-Tolerant CeratozamiaCeratozamiaCeratozamiaCeratozamia SpeciesSpeciesSpeciesSpecies ☻Shade Loving ☻Cold TolerTolerantant ▲Filtered/Coastal Sun 16 named + several undescribed species Native to Mexico, Guatemala & Belize Name originates from Greek ceratos (horned), and azaniae, (pine cone) Pinnate (feather-shaped) leaves, lacking a midrib, and horned, spiny cones Shiny, darker green leaves arching or upright, often emerging red or brown Less “formal” looking than other cycads Prefer Shade ½ - ¾ day, or afternoon shade Generally cold-tolerant CeratozamiaCeratozamia ---- SuggestedSuggested SpeciesSpecies ☻Shade Loving ☻Cold TolerTolerantant ▲Filtered/Coastal Sun Ceratozamia mexicana Tropical looking but cold-tolerant, native to dry mountainous areas in the Sierra Madre Mountains (Mexican Rockies). Landscape specimen works well with water features, due to arching habit. Prefers shade, modest height, with a spread of up to 10 feet. Trunk grows to 2 feet tall. Leaflets can be narrow or wider (0.75-2 inches). CeratozamiaCeratozamia ---- SuggestedSuggested SpeciesSpecies ☻Shade Loving ☻Cold TolerTolerantant ▲Filtered/Coastal Sun Ceratozamia latifolia Rare Ceratozamia named for its broad leaflets. Native to cloud forests of the Sierra Madre mountains of Mexico, underneath oak trees. Emergent trunk grows to 1 foot tall, 8 inches in diameter. New leaves emerge bronze, red or chocolate brown, hardening off to bright green, semiglossy, and grow to 6 feet long. They are flat lance-shaped, asymmetric, and are broadest above middle, growing to 10 inches long and 2 inches wide. -

Crime, Culture and Collecting: the Illicit Cycad Market in South Africa
CRIME, CULTURE AND COLLECTING: THE ILLICIT CYCAD MARKET IN SOUTH AFRICA BY JONAS SØRFLATEN TORGERSEN SUPERVISED BY Town PROFESSOR MARK SHAW Cape of THESIS Submitted in partial fulfilment of the requirements for the degree of Master of Philosophy in University Criminology, Law and Society in the Faculty of Law at the University of Cape Town, 2017 Cape Town, South Africa The copyright of this thesis vests in the author. No quotation from it or information derived from it is to be published without full acknowledgement of the source. The thesis is to be used for private study or non- commercial research purposes only. Published by the University of Cape Town (UCT) in terms of the non-exclusive license granted to UCT by the author. University of Cape Town CRIME, CULTURE AND COLLECTING: THE ILLICIT CYCAD MARKET IN SOUTH AFRICA Abstract It is widely accepted that illicit markets are driven by specific contextual factors that determine their nature and scope. Two points in particular have not been explored in the literature on wildlife crime. First, while illicit markets around commodities such as drugs and weapons are fuelled by consumers arguably in need of, or addicted to, the product, the desires of buyers that shape wildlife markets are often shaped by cultural norms which may seem irrational to outsiders. Second, given that wildlife markets are seldom as stringently regulated as those in respect of drugs, weapons or other commodities, the nature of the criminal enterprises that source, move and sell the products are possibly very different. The study examines these two factors – the culture of markets and the degree of criminal enterprise or organisation within them – through a case study of a largely unexamined environmental crime market in South Africa, that of rare cycad plants. -

Inducing Sex Change and Organogenesis from Tissue Culture
114 SouthAfrican Journal of Science98, March/April2002 Gommentary reveals slight heterochromatin differ- lnducing sex change and ences,"'" which is itself due to differential methylation."''n Therefore, it is distinctly organogenesis from tissue possible that methylation controls sex determination. culture in the endangered African Methylation and accompanying hetero- chromatin can be removed by various cycad Encephalartos woodii f actors - such as temper attJte,'5''6 lighft ,27 osmotic stress,28or hormones2e-31- result- (Gycadales, Zamiaceael ing in sex change.3''33Sex change occurs only in organisms that have (virtually) indistinguishable sex chromosomes, indi- Root Gorelicku" and Roy Osborneb cating that incipient sex chromosomes are formed by slight differences in methylation. Differential methylation is I F INCIPIENT SEX CHROMOSOME DIFFEREN- Fourth, if backcrossing is the only realistic evolutionarily the first difference between I tiutiott is caused by differential methy- approach to conservation, then it is pref- females and males3nand is the likely cause I lation between females and males, then erableto use E.woodiiasthe female parent of reported sex changes in cycads. methylating or demethylating cytosine because of maternal inheritance of nucleotides may induce sex change. chloroplast genomes. Fifth, induced sex Methylation may also stimulate regeneration Application of theory to sex change of roots and shoots from tissue culture callus change may assist in the conservation of in cycads and increase genetic variation via greater