Supplemental Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Transcriptional Regulation of RKIP in Prostate Cancer Progression
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Sciences Transcriptional Regulation of RKIP in Prostate Cancer Progression Submitted by: Sandra Marie Beach In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Sciences Examination Committee Major Advisor: Kam Yeung, Ph.D. Academic William Maltese, Ph.D. Advisory Committee: Sonia Najjar, Ph.D. Han-Fei Ding, M.D., Ph.D. Manohar Ratnam, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: May 16, 2007 Transcriptional Regulation of RKIP in Prostate Cancer Progression Sandra Beach University of Toledo ACKNOWLDEGMENTS I thank my major advisor, Dr. Kam Yeung, for the opportunity to pursue my degree in his laboratory. I am also indebted to my advisory committee members past and present, Drs. Sonia Najjar, Han-Fei Ding, Manohar Ratnam, James Trempe, and Douglas Pittman for generously and judiciously guiding my studies and sharing reagents and equipment. I owe extended thanks to Dr. William Maltese as a committee member and chairman of my department for supporting my degree progress. The entire Department of Biochemistry and Cancer Biology has been most kind and helpful to me. Drs. Roy Collaco and Hong-Juan Cui have shared their excellent technical and practical advice with me throughout my studies. I thank members of the Yeung laboratory, Dr. Sungdae Park, Hui Hui Tang, Miranda Yeung for their support and collegiality. The data mining studies herein would not have been possible without the helpful advice of Dr. Robert Trumbly. I am also grateful for the exceptional assistance and shared microarray data of Dr. -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

(12) United States Patent (10) Patent No.: US 9,005,612 B2 Ledbetter Et Al
US009005612B2 (12) United States Patent (10) Patent No.: US 9,005,612 B2 Ledbetter et al. (45) Date of Patent: *Apr. 14, 2015 (54) BINDING DOMAIN-IMMUNOGLOBULIN 5,455,030 A 10/1995 Ladner et al. FUSION PROTEINS $599. A 3: BestInsley et al. al. (75) Inventors: Jeffrey A. Ledbetter, Seattle, WA (US); 5,530,101 A 6/1996 Queen et al. Martha S. Hayden-Ledbetter, Seattle, 5,580,756. A 12/1996 Linsley et al. WA (US) 5,595,7215,585,089 A 1431/1997 SAS,Kaminski et al.1 (73) Assignee: Emergent Product Development 5,597,707 A 1/1997 Marken et al. Seattle, LLC, Seattle, WA (US) 3.W - A 3.87 EaCee ea. (*) Notice: Subject to any disclaimer, the term of this 5,645,835 A 7/1997 Fell, Jr. et al. patent is extended or adjusted under 35 $22.9 A ck 8. 3. Earl 530,387.1 U.S.C. 154(b) by 0 days. 5,693,762.w A 12/1997 QueenOCC et Cal. al. ............ This patent is Subject to a terminal dis- 5,709,859 A 1/1998 Aruffo et al. claimer 5,714,147 A 2, 1998 Capon et al. 5,721, 108 A 2f1998 Robinson et al. (21) Appl. No.: 13/451,641 5,736,137 A 4, 1998 Anderson et al. 5,770,197 A 6/1998 Linsley et al. (22) Filed: Apr. 20, 2012 5,773.253 A 6/1998 Linsley et al. O O 5,776.456 A 7/1998 Anderson et al. (65) Prior Publication Data 5,795,572 A 8/1998 Diegel et al. -

Atpase, Na+/K+ Transporting, Alpha 3 Polypeptide Homologous to 3'UTR
HUGO ID Name Nalm-6 TOM-1 Reh Karpas-422 DoHH -2 SU-DHL-5 Namalwa DG-75 Ramos Raji BEL EHEB BONNA-12 L-428 DEL BCP-1 BC-3 BCBL-1 JSC-1 PEL-SY HBL-6 DS-1 RPMI-8226 NCI-H929 L-363 SK-MM-2 ATP1A3 ATPase, Na+/K+ transporting, alpha 3 polypeptide CD24 homologous to 3'UTR of human CD24 gene ABCC5 multidrug resistance-associated protein (MRP5) CD72 CD72 antigen TCL1A Tcell leukemia/lymphoma 1 ITGB2 Integrin, beta 2 (antigen CD18 (p95)) ? nuclear ribonucleoprotein particle (hnRNP) SGT1 suppressor of G2 allele of skp1 homolog DNMT 1 DNA (cytosine-5-)-methyltransferase 1 GALE UDP-Galactose 4 epimerase (GALE) HADHSC L-3-hydroxyacyl-CoA dehydrogenase LIG4 DNA ligase IV LIG1 Ligase I, DNA, ATP-dependent CEBPG CCAA T/enhancer binding protein (C/EBP), gamma DCK Deoxycytidine kinase TCEA1 TRANSCRIPTION ELONGATION FACTOR S-II TCN 1 TRANSCOBALAMIN I PRECURSOR POLA2 DNA polymerase alpha subunit CCNG2 cyclin G2 RNPC1 Finkel-Biskis-Reilly murine sarcoma virus; Human seb4D RNPC1 Finkel-Biskis-Reilly murine sarcoma virus; Human seb4D DGKD Diacylglycerol kinase delta KIAA0220 Polycystic kidney disease protein 1 KIAA0220 calcium-dependent group X phospholipase A2 KIAA0220 calcium-dependent group X phospholipase A2 ALDH5A1 NAD+-dependent succinate-semialdehyde dehydrogenase CCNG2 Polycystic kidney disease 1 (autosomal dominant) PDCD4 nuclear antigen H731-like protein SSH3BP1 eps8 binding protein e3B1 MAP4K2 B lymphocyte serine/threonine protein kinase (GC kinase) MAPRE2 novel T-cell activation protein ZNFN1A Ikaros/LyF-1 homolog (hIk-1) FLJ22624 clone 23799 KIAA0355 -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Development of a New Screening System for the Identification of RNF43-Related Genes and Characterisation of Other PA-RING Family Members
Development of a new screening system for the identification of RNF43-related genes and characterisation of other PA-RING family members Alessandra Merenda University of Cambridge Department of Genetics This dissertation is submitted for the degree of Doctor of Philosophy Darwin College April 2017 ii | P a g e This dissertation is dedicated to my exemplary grandfather. iv | P a g e Declaration I hereby declare that, except where specific reference is made to the work of others, the contents of this dissertation are original and they are the result of my own work. This dissertation contains nothing which is the outcome of work done in collaboration with others, except as specified in the text and under “External contributions”. It is not substantially the same as any that I have submitted, or, is being concurrently submitted for a degree or diploma or other qualification at the University of Cambridge or any other University or similar institution. This dissertation does not exceed the prescribed word limit of 60,000 words excluding bibliography, figures and appendices. Alessandra Merenda April 2017 v | P a g e External contributions The details about external contributions to the work presented in this thesis are as follows: Dr. Ingrid Jordens (UMC) and Dr. Cara Jamieson (UMC) performed the pulse chase experiments with SNAP-Frizzled5 (Figure 5.2 and Figure 5.3). Dr. Jihoon Kim (University of Cambridge) performed FZD5 western blot and FACS analysis, ubiquitination assay and co-immunoprecipitation (Figures 5.4-5.7). vi | P a g e vii | P a g e Acknowledgments As this intense journey comes to an end, many are the people I feel like thanking for making it worth undertaking. -
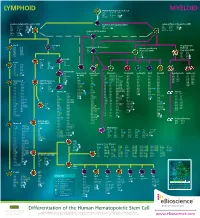
Lymphoid Myeloid
LYMPHOID Human Hematopoietic Stem Cell MYELOID CD34 CD117 (c-kit) CD338 CD38low/neg CD133 linneg CD59 CD135 (Flt3) GATA2 CD90 (Thy1) CD164 TdT Common Lymphoid Progenitor (CLP) Multi-Potent Progenitor (MPP) Common Myeloid Progenitor (CMP) low neg CD33 CD123low CD174 CD7 CD117 (c-kit) C/EBPα CD34 linneg neg CD34 CD131 Ikaros CD10 CD127 GATA2 CD135 (Flt3) TdT CD24neg CD135 (Flt3) GATA3 CD45RA CD164 PU.1 CD34 CD164 PU.1high Common DC Progenitor CD117 (c-kit) CD173 CD38 HLA-DR TdT CD45RA Aiolos CD11c dim CD90low c-mybhigh CD33 Pro-B Pre-NK/T Megakaryocyte- CD10 CD124 CD5 Conventional DC Precursor Erythroid CD19 CD164 CD34 CD11c Granulocyte-Myeloid CD20 CD252 CD14neg Progenitor (GMP) Progenitor (MEP) CD22 CD268 CD33dim CD33 CD131 CD34neg CD275 CD34 CD45RA CD34 CD164 CD72 HLA-DR CD36 FOG-1 CD74 CD123 PU.1 CD110 GATA1 CD123high GATA2 Pre-T Pro-NK CD7 Pre-B CD1a CD28 CD10neg CD2 CD34 neg CD10 CD124 CD34 CD3 CD45 CD117 (c-kit) CD19 CD164 CD5 CD127 (IL-7Rα) CD20 CD252 CD7high NOTCH1 CD22 CD268 CD34neg CD275 CD72 HLA-DR Immature NK CD74 IgM CD34neg Plasmacytoid Conventional Monocyte Neutrophil Eosinophil Mast Basophil Megakaryocyte Erythrocyte CD79a Pax5 CD94neg DC (pDC) DC (cDC) CD4low CD85a (ILT5) CD171 CD10 CD9 CD9 CD9 CD41 CD51 CD35 CD236 CD117 (c-kit) CD9 CD85d (ILT4) CD172a (SIRPα) CD15 CD11b CD11b CD11a CD42a CD61 CD44 CD236R CD4 CD1c CD122 CD11b CD85h (ILT1) CD172b CD16b CD15 CD15 CD11b CD42b CD110 CD123 CD238 CD11c neg CD1d CD161 CD11c CD85j (ILT2) CD180 CD17 CD24 CD24 CD13 CD42c CD112 CD173 CD239 CD45RA CD2 Ets-1 CD13 -

Human CD Marker Chart Reviewed by HLDA1 Bdbiosciences.Com/Cdmarkers
BD Biosciences Human CD Marker Chart Reviewed by HLDA1 bdbiosciences.com/cdmarkers 23-12399-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1a R4, T6, Leu6, HTA1 b-2-Microglobulin, CD74 + + + – + – – – CD93 C1QR1,C1qRP, MXRA4, C1qR(P), Dj737e23.1, GR11 – – – – – + + – – + – CD220 Insulin receptor (INSR), IR Insulin, IGF-2 + + + + + + + + + Insulin-like growth factor 1 receptor (IGF1R), IGF-1R, type I IGF receptor (IGF-IR), CD1b R1, T6m Leu6 b-2-Microglobulin + + + – + – – – CD94 KLRD1, Kp43 HLA class I, NKG2-A, p39 + – + – – – – – – CD221 Insulin-like growth factor 1 (IGF-I), IGF-II, Insulin JTK13 + + + + + + + + + CD1c M241, R7, T6, Leu6, BDCA1 b-2-Microglobulin + + + – + – – – CD178, FASLG, APO-1, FAS, TNFRSF6, CD95L, APT1LG1, APT1, FAS1, FASTM, CD95 CD178 (Fas ligand) + + + + + – – IGF-II, TGF-b latency-associated peptide (LAP), Proliferin, Prorenin, Plasminogen, ALPS1A, TNFSF6, FASL Cation-independent mannose-6-phosphate receptor (M6P-R, CIM6PR, CIMPR, CI- CD1d R3G1, R3 b-2-Microglobulin, MHC II CD222 Leukemia -

LECTURE 07 Immune Cells Surface Receptors and Their Functions
LECTURE: 07 Title: IMMUNE CELL SURFACE RECEPTORS AND THEIR FUNCTIONS LEARNING OBJECTIVES: The student should be able to: • The chemical nature of the cellular surface receptors. • Define the location of the cellular receptors. • Enumerate the different given terms for the word "receptor". • Define the term "CD" marker. • Describe the location of the immune receptors. • Classify the cellular receptors according to the information they offer. • Classify the cellular receptors according to their families. • Enumerate some common immunoglobulin receptors. • Enumerate some common complement receptors. • Enumerate some adhesion molecules receptors. • Enumerate the main functions of the immune cell receptors. • Realize the importance of the immune receptors. LECTURE REFRENCE: 1. TEXTBOOK: ROITT, BROSTOFF, MALE IMMUNOLOGY. 6th edition. pp. 29, 143, 76, 77, 78, 221, 325. 2. TEXTBOOK: ABUL K. ABBAS. ANDREW H. LICHTMAN. CELLULAR AND MOLECULAR IMMUNOLOGY. 5TH EDITION. Chapter. 6 .pg 105. Chapter 4. pg 65- 80. 1 Lymphocytes can be identified by characteristic markers Lymphocytes and other leukocytes express different surface molecules which can be used for distinguishing cell population. A systemic nomenclature called the CD system has been developed, in which the term CD refers to (cluster of designation). These different cell surface molecules are detected with specific monoclonal antibodies, each of these molecules are given CD number. Molecular markers can be further defined according to the information they offer about the cell. For example; A. Lineage markers are exclusive to a particular cell line, e.g., CD3, which is found only on T cells. B. Maturation markers are transiently expressed during cell differentiation, e.g., CD1, which is only found on cells developing in thymus and is not present on peripheral T cells.