New Phylogenomic Data Support the Monophyly of Lophophorata and An
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Platyhelminthes, Nemertea, and "Aschelminthes" - A
BIOLOGICAL SCIENCE FUNDAMENTALS AND SYSTEMATICS – Vol. III - Platyhelminthes, Nemertea, and "Aschelminthes" - A. Schmidt-Rhaesa PLATYHELMINTHES, NEMERTEA, AND “ASCHELMINTHES” A. Schmidt-Rhaesa University of Bielefeld, Germany Keywords: Platyhelminthes, Nemertea, Gnathifera, Gnathostomulida, Micrognathozoa, Rotifera, Acanthocephala, Cycliophora, Nemathelminthes, Gastrotricha, Nematoda, Nematomorpha, Priapulida, Kinorhyncha, Loricifera Contents 1. Introduction 2. General Morphology 3. Platyhelminthes, the Flatworms 4. Nemertea (Nemertini), the Ribbon Worms 5. “Aschelminthes” 5.1. Gnathifera 5.1.1. Gnathostomulida 5.1.2. Micrognathozoa (Limnognathia maerski) 5.1.3. Rotifera 5.1.4. Acanthocephala 5.1.5. Cycliophora (Symbion pandora) 5.2. Nemathelminthes 5.2.1. Gastrotricha 5.2.2. Nematoda, the Roundworms 5.2.3. Nematomorpha, the Horsehair Worms 5.2.4. Priapulida 5.2.5. Kinorhyncha 5.2.6. Loricifera Acknowledgements Glossary Bibliography Biographical Sketch Summary UNESCO – EOLSS This chapter provides information on several basal bilaterian groups: flatworms, nemerteans, Gnathifera,SAMPLE and Nemathelminthes. CHAPTERS These include species-rich taxa such as Nematoda and Platyhelminthes, and as taxa with few or even only one species, such as Micrognathozoa (Limnognathia maerski) and Cycliophora (Symbion pandora). All Acanthocephala and subgroups of Platyhelminthes and Nematoda, are parasites that often exhibit complex life cycles. Most of the taxa described are marine, but some have also invaded freshwater or the terrestrial environment. “Aschelminthes” are not a natural group, instead, two taxa have been recognized that were earlier summarized under this name. Gnathifera include taxa with a conspicuous jaw apparatus such as Gnathostomulida, Micrognathozoa, and Rotifera. Although they do not possess a jaw apparatus, Acanthocephala also belong to Gnathifera due to their epidermal structure. ©Encyclopedia of Life Support Systems (EOLSS) BIOLOGICAL SCIENCE FUNDAMENTALS AND SYSTEMATICS – Vol. -

Spermiogenesis in the Acoel Symsagittifera Roscoffensis: Nucleus-Plasma
bioRxiv preprint doi: https://doi.org/10.1101/828251; this version posted November 1, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Spermiogenesis in the acoel Symsagittifera roscoffensis: nucleus-plasma membrane contact sites and microtubules. Matthew J. Hayes1, Anne-C. Zakrzewski2, Tim P. Levine1 and Maximilian J. Telford2 1University College London Institute of Ophthalmology, 11-43 Bath Street, London EC1V 9EL, UK 2 Centre for Life’s Origins and Evolution, Department of Genetics, Evolution and Environment, University College London, Gower Street, London, WC1E 6BT, UK Running title: Contact sites and microtubule rearrangements in spermiogenesis Summary sentence: During spermiogenesis in the acoel flatworm Symsagittifera roscoffensis, two previously unidentified contact sites contribute to the structure of the mature spermatozoon and the axonemal structures show direct continuity between doublet and dense core microtubules. Keywords: Contact-site, acrosome, gamete biology, spermatid, spermatogenesis, spermiogenesis, acoel. bioRxiv preprint doi: https://doi.org/10.1101/828251; this version posted November 1, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Symsagittifera roscoffensis is a small marine worm found in the intertidal zone of sandy beaches around the European shores of the Atlantic. S. roscoffensis is a member of the Acoelomorpha, a group of flatworms formerly classified with the Platyhelminthes, but now recognised as Xenacoelomorpha, a separate phylum of disputed affinity. -

List of Marine Alien and Invasive Species
Table 1: The list of 96 marine alien and invasive species recorded along the coastline of South Africa. Phylum Class Taxon Status Common name Natural Range ANNELIDA Polychaeta Alitta succinea Invasive pile worm or clam worm Atlantic coast ANNELIDA Polychaeta Boccardia proboscidea Invasive Shell worm Northern Pacific ANNELIDA Polychaeta Dodecaceria fewkesi Alien Black coral worm Pacific Northern America ANNELIDA Polychaeta Ficopomatus enigmaticus Invasive Estuarine tubeworm Australia ANNELIDA Polychaeta Janua pagenstecheri Alien N/A Europe ANNELIDA Polychaeta Neodexiospira brasiliensis Invasive A tubeworm West Indies, Brazil ANNELIDA Polychaeta Polydora websteri Alien oyster mudworm N/A ANNELIDA Polychaeta Polydora hoplura Invasive Mud worm Europe, Mediterranean ANNELIDA Polychaeta Simplaria pseudomilitaris Alien N/A Europe BRACHIOPODA Lingulata Discinisca tenuis Invasive Disc lamp shell Namibian Coast BRYOZOA Gymnolaemata Virididentula dentata Invasive Blue dentate moss animal Indo-Pacific BRYOZOA Gymnolaemata Bugulina flabellata Invasive N/A N/A BRYOZOA Gymnolaemata Bugula neritina Invasive Purple dentate mos animal N/A BRYOZOA Gymnolaemata Conopeum seurati Invasive N/A Europe BRYOZOA Gymnolaemata Cryptosula pallasiana Invasive N/A Europe BRYOZOA Gymnolaemata Watersipora subtorquata Invasive Red-rust bryozoan Caribbean CHLOROPHYTA Ulvophyceae Cladophora prolifera Invasive N/A N/A CHLOROPHYTA Ulvophyceae Codium fragile Invasive green sea fingers Korea CHORDATA Actinopterygii Cyprinus carpio Invasive Common carp Asia CHORDATA Ascidiacea -

Cribrilina Mutabilisn. Sp., an Eelgrass-Associated Bryozoan (Gymnolaemata: Cheilostomata) with Large Variationin Title Zooid Morphology Related to Life History
Cribrilina mutabilisn. sp., an Eelgrass-Associated Bryozoan (Gymnolaemata: Cheilostomata) with Large Variationin Title Zooid Morphology Related to Life History Author(s) Ito, Minako; Onishi, Takumi; Dick, Matthew H. Zoological Science, 32(5), 485-497 Citation https://doi.org/10.2108/zs150079 Issue Date 2015-10 Doc URL http://hdl.handle.net/2115/62926 Type article File Information ZS32-5 485-497.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP ZOOLOGICAL SCIENCE 32: 485–497 (2015) © 2015 Zoological Society of Japan Cribrilina mutabilis n. sp., an Eelgrass-Associated Bryozoan (Gymnolaemata: Cheilostomata) with Large Variation in Zooid Morphology Related to Life History Minako Ito1, Takumi Onishi2, and Matthew H. Dick2* 1Graduate School of Environmental Science, Hokkaido University, Aikappu 1, Akkeshi-cho, Akkeshi-gun 088-1113, Japan 2Department of Natural History Sciences, Faculty of Science, Hokkaido University, N10 W8, Sapporo 060-0810, Japan We describe the cribrimorph cheilostome bryozoan Cribrilina mutabilis n. sp., which we detected as an epibiont on eelgrass (Zostera marina) at Akkeshi, Hokkaido, northern Japan. This species shows three distinct zooid types during summer: the R (rib), I (intermediate), and S (shield) types. Evidence indicates that zooids commit to development as a given type, rather than transform from one type to another with age. Differences in the frontal spinocyst among the types appear to be mediated by a simple developmental mechanism, acceleration or retardation in the production of lateral costal fusions as the costae elongate during ontogeny. Colonies of all three types were identical, or nearly so, in partial nucleotide sequences of the mitochondrial COI gene (555–631 bp), suggesting that they represent a single species. -

Leafy Bryozoans and Other Bryozoan Species in General Play an Important Role in Marine Ecosystems
These wash up on the shoreline looking like Flustra foliacea clumps of seaweed. Class: Gymnolaemata Order: Cheilostomata Family: Flustridae Genus: Flustra Distribution Flustra foliacea has a wide It is common to the coastal areas of northern Europe especially distribution in the North in the North Sea. Countries include Britain, Ireland, Belgium, Atlantic Ocean, on both the Netherlands, and France. It does not continue any further south European and American than northern Spain. In Canada it is in Nova Scotian waters, sides. including the Bay of Fundy and the Minas Basin. Habitat It most frequently occurs between 10-20 m water depths. It is This is a cold water species. typically found on the upper faces of moderately wave-exposed It prefers high salinity bedrock or boulders subjected to moderately strong tidal waters, but can also found streams. These rocky patches may be interspersed with gravelly in areas with lower salinity. sand patches, causing a scouring effect. Most Bryozoans live in It occupies sublittoral salt water, and of the 20 or so freshwater species found in North (below low tide) areas. America, most are found in warm-water regions attached to plants, logs, rocks and other firm substrates. Food This species is an active This is a colonial animal composed of various types of zooids. A suspension feeder. They zooid is a single animal that is part of the colony. The basic consume phytoplankton, zooids are the feeding ones, called the autozooid. Each of these detritus, and dissolved has a mouth and a feeding structure, the lophophore, which is organic matter. -

Xenacoelomorpha's Significance for Understanding Bilaterian Evolution
Available online at www.sciencedirect.com ScienceDirect Xenacoelomorpha’s significance for understanding bilaterian evolution Andreas Hejnol and Kevin Pang The Xenacoelomorpha, with its phylogenetic position as sister biology models are the fruitfly Drosophila melanogaster and group of the Nephrozoa (Protostomia + Deuterostomia), plays the nematode Caenorhabditis elegans, in which basic prin- a key-role in understanding the evolution of bilaterian cell types ciples of developmental processes have been studied in and organ systems. Current studies of the morphological and great detail. It might be because the field of evolutionary developmental diversity of this group allow us to trace the developmental biology — EvoDevo — has its origin in evolution of different organ systems within the group and to developmental biology and not evolutionary biology that reconstruct characters of the most recent common ancestor of species under investigation are often called ‘model spe- Xenacoelomorpha. The disparity of the clade shows that there cies’. Criteria for selected representative species are cannot be a single xenacoelomorph ‘model’ species and primarily the ease of access to collected material and strategic sampling is essential for understanding the evolution their ability to be cultivated in the lab [1]. In some cases, of major traits. With this strategy, fundamental insights into the a supposedly larger number of ancestral characters or a evolution of molecular mechanisms and their role in shaping dominant role in ecosystems have played an additional animal organ systems can be expected in the near future. role in selecting model species. These arguments were Address used to attract sufficient funding for genome sequencing Sars International Centre for Marine Molecular Biology, University of and developmental studies that are cost-intensive inves- Bergen, Thormøhlensgate 55, 5008 Bergen, Norway tigations. -

"Phoronida". In: Encyclopedia of Life Science
Phoronida Introductory article Christian C Emig, Centre d’Oce´anologie de Marseille CNRS, Marseille, France Article Contents . Basic Design The Phoronida, divided into two genera and 10 species, is a small marine group, which . Diversity and Lifestyles belongs to the phylum Lophophorata. Fossil History and Phylogeny Basic Design complexity of the lophophore from an oval to a helicoidal, The Phoronida is an exclusively marine group with a sessile through a horseshoe and spiral shape. This is related to an vermiform body enclosed in a tube (Figure 1) The body is increase in the number of tentacles, which is proportional composed of three distinct parts (prosome, mesosome and to the general body size. metasome), each containing its own coelomic cavity. The The metasome (or trunk) is slender and cylindrical, with prosome forms the epistome, a fold overhanging the mouth a bulb-like posterior end (or ampulla) that anchors the dorsally. The mesosome bears the lophophore, with the body in the rear end of the tube. It is separated from the rest mouth lying between its two rows of tentacles. The of the body by the diaphragm, a thick transverse septum lophophore is a terminal, bilaterally symmetrical, tentacle located behind the lophophore. crown, each tentacle having complex arrays of cilia for The digestive tract is U-shaped, bringing the anus close filter-feeding. Lophophore shape is a fairly constant to the mouth. The descending branch is divided into a short feature within each species, and there is an increasing oesophagus, followed by a long prestomach, then a stomach surrounded by a blood plexus. -

A Phylogenetic Analysis of Myosin Heavy Chain Type II Sequences Corroborates That Acoela and Nemertodermatida Are Basal Bilaterians
A phylogenetic analysis of myosin heavy chain type II sequences corroborates that Acoela and Nemertodermatida are basal bilaterians I. Ruiz-Trillo*, J. Paps*, M. Loukota†, C. Ribera†, U. Jondelius‡, J. Bagun˜ a` *, and M. Riutort*§ *Departament de Gene`tica and †Departament de Biologia Animal, Universitat Barcelona, Av. Diagonal, 645 08028 Barcelona, Spain; and ‡Evolutionary Biology Centre, University Uppsala, Norbyva¨gen 18D, 752 36 Uppsala, Sweden Communicated by James W. Valentine, University of California, Berkeley, CA, June 28, 2002 (received for review April 15, 2002) Bilateria are currently subdivided into three superclades: Deuter- across all bilaterians. However different, this scheme also suited ostomia, Ecdysozoa, and Lophotrochozoa. Within this new taxo- alternative hypotheses such as the ‘‘set-aside cells’’ theory (13, nomic frame, acoelomate Platyhelminthes, for a long time held to 14) and the colonial ancestor theory (15). be basal bilaterians, are now considered spiralian lophotrochozo- This new status quo was soon questioned. A SSU-based study ans. However, recent 18S rDNA [small subunit (SSU)] analyses have using a large set of Platyhelminthes acoels and other metazoans shown Platyhelminthes to be polyphyletic with two of its orders, showed acoels to be the extant earliest branching bilaterians the Acoela and the Nemertodermatida, as the earliest extant (16), turning Platyhelminthes into a polyphyletic group. In bilaterians. To corroborate such position and avoid the criticisms of addition, Jenner (17) noted that phylogenies put forward to back saturation and long-branch effects thrown on the SSU molecule, the new metazoan molecular trees (10–12, 18) were incomplete we have searched for independent molecular data bearing good and heavily pruned. -

New and Known Nemertodermatida (Platyhelminthes-Acoelomorpha)
Belg. J. Zoo!. - Volume 128 (1998) - issue 1 - pages 55-92 - Brussels 1998 Received: 18 February 1998 NEW AND KNOWN NEMERTODERMATIDA (PLATYHELMINTHES-ACOELOMORPHA)... -A REVISION - WOLFGANG STERRER Bermuda Natural History Museum, Flatts FLBX, Bermuda e-mail [email protected] Abstract. Described in 1930-31 by Steinbiick who considered it the most primitive bilaterian, the turbellarian genus Nemertoderma is known for its rote in platyhelminth phylogeny as much as for its muddled taxonomy. On the basis of material collected in the Mediterranean, Atlantic and Pacifie Oceans sin ce 1964 this paper re-diagnoses the known 4 genera and 7 species (Nemertoderma bathycola Steinbiick, 1930-31 ; N. westbladi Steinbiick, 1938; N. psammicola Sterrer, 1970 (syn. N. rubra Faubel, 1976); Meara stichopi Westblad, 1949; Meara sp. (see SMITH et al., 1994); Nemertinoides elongatus Riser, 1987 ; and Flagellophora apelti Fau bel & Diirjes, 1978), describes one new genus with 2 new species (Ascoparia neglecta n. g., n. sp. and A. secunda n. sp.), and pro vides observations from living material on morphological variability, body size vs . reproductive state, statocyst structure and statolith variability, and sperm morphology and dimorphism. The paper concludes with diagnoses for the known taxa of Nemertodermatida, including the new family Ascopariidae. Key words: Platyhelminthes, free-living, marine; systematics, new species. INTRODUCTION ln 1930-31 Otto STEfNBOCK described Nemertoderma bathycola from a single tiny worm whicb he and Erich Reisinger bad dredged from a muddy bottom, at 300-400 rn depth, off Greenland. Nemertoderma caused a small sensation, not only because Steinbock, a meticulous observer, was also an assertive character (who liked to express himself double-spaced, with exclamation marks added) but because Nemertoderma was indeed unusual. -
S I Section 4
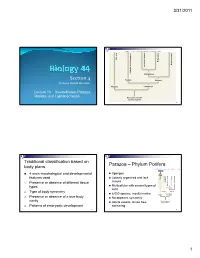
3/31/2011 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Porifera Ecdysozoa Deuterostomia Lophotrochozoa Cnidaria and Ctenophora Cnidaria and Protostomia SSiection 4 Radiata Bilateria Professor Donald McFarlane Parazoa Eumetazoa Lecture 13 Invertebrates: Parazoa, Radiata, and Lophotrochozoa Ancestral colonial choanoflagellate 2 Traditional classification based on Parazoa – Phylum Porifera body plans Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Parazoa 4 main morphological and developmental Sponges features used Loosely organized and lack Porifera Ecdysozoa Cnidaria and Ctenophora tissues euterostomia photrochozoa D 1. o Presence or absence of different tissue L Multicellular with several types of types Protostomia cells 2. Type of body symmetry Radiata Bilateria 8,000 species, mostly marine Parazoa Eumetazoa 3. Presence or absence of a true body No apparent symmetry cavity Ancestral colonial Adults sessile, larvae free- choanoflagellate 4. Patterns of embryonic development swimming 3 4 1 3/31/2011 Water drawn through pores (ostia) into spongocoel Flows out through osculum Reproduce Choanocytes line spongocoel Sexually Most hermapppgggphrodites producing eggs and sperm Trap and eat small particles and plankton Gametes are derived from amoebocytes or Mesohyl between choanocytes and choanocytes epithelial cells Asexually Amoebocytes absorb food from choanocytes, Small fragment or bud may detach and form a new digest it, and carry -

Recolonization of Freshwater Ecosystems Inferred from Phylogenetic Relationships Nikola Koleti�C1, Maja Novosel2, Nives Rajevi�C2 & Damjan Franjevi�C2
Bryozoans are returning home: recolonization of freshwater ecosystems inferred from phylogenetic relationships Nikola Koletic1, Maja Novosel2, Nives Rajevic2 & Damjan Franjevic2 1Institute for Research and Development of Sustainable Ecosystems, Jagodno 100a, 10410 Velika Gorica, Croatia 2Department of Biology, Faculty of Science, University of Zagreb, Rooseveltov trg 6, 10000 Zagreb, Croatia Keywords Abstract COI, Gymnolaemata, ITS2, Phylactolaemata, rRNA genes. Bryozoans are aquatic invertebrates that inhabit all types of aquatic ecosystems. They are small animals that form large colonies by asexual budding. Colonies Correspondence can reach the size of several tens of centimeters, while individual units within a Damjan Franjevic, Department of Biology, colony are the size of a few millimeters. Each individual within a colony works Faculty of Science, University of Zagreb, as a separate zooid and is genetically identical to each other individual within Rooseveltov trg 6, 10000 Zagreb, Croatia. the same colony. Most freshwater species of bryozoans belong to the Phylacto- Tel: +385 1 48 77 757; Fax: +385 1 48 26 260; laemata class, while several species that tolerate brackish water belong to the E-mail: [email protected] Gymnolaemata class. Tissue samples for this study were collected in the rivers of Adriatic and Danube basin and in the wetland areas in the continental part Funding Information of Croatia (Europe). Freshwater and brackish taxons of bryozoans were geneti- This research was supported by Adris cally analyzed for the purpose of creating phylogenetic relationships between foundation project: “The genetic freshwater and brackish taxons of the Phylactolaemata and Gymnolaemata clas- identification of Croatian autochthonous ses and determining the role of brackish species in colonizing freshwater and species”. -

Frontiers in Zoology Biomed Central
Frontiers in Zoology BioMed Central Research Open Access Myogenesis in the basal bilaterian Symsagittifera roscoffensis (Acoela) Henrike Semmler*1, Xavier Bailly2 and Andreas Wanninger1 Address: 1University of Copenhagen, Department of Biology, Research Group for Comparative Zoology, Universitetsparken 15, DK-2100 Copenhagen Ø, Denmark and 2Station Station Biologique de Roscoff, Place Georges Teissier BP74, F-29682 Roscoff Cedex, France Email: Henrike Semmler* - [email protected]; Xavier Bailly - [email protected]; Andreas Wanninger - [email protected] * Corresponding author Published: 19 September 2008 Received: 5 May 2008 Accepted: 19 September 2008 Frontiers in Zoology 2008, 5:14 doi:10.1186/1742-9994-5-14 This article is available from: http://www.frontiersinzoology.com/content/5/1/14 © 2008 Semmler et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: In order to increase the weak database concerning the organogenesis of Acoela – a clade regarded by many as the earliest extant offshoot of Bilateria and thus of particular interest for studies concerning the evolution of animal bodyplans – we analyzed the development of the musculature of Symsagittifera roscoffensis using F-actin labelling, confocal laserscanning microscopy, and 3D reconstruction software. Results: At 40% of development between egg deposition and hatching short subepidermal fibres form. Muscle fibre development in the anterior body half precedes myogenesis in the posterior half. At 42% of development a grid of outer circular and inner longitudinal muscles is present in the bodywall.