Molecular Identification, Occurrence
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

180 Kb Herrera
CALENDARIOS DE BECA UNIVERSAL Y PLANILLA GLOBAL PRIMERA ENTREGA 2019 REGIONAL DE HERRERA LUNES 11 , DE MARZO DE 2019 # DISTRITOS CENTRO EDUCATIVOS CENTRO DE PAGO 1 CHITRÉ ESCUELA ENEIDA M., DE CASTILLERO 2 CHITRÉ ESCUELA BOCA DE PARITA 3 CHITRÉ IPHE CHITRÉ 4 CHITRÉ ESCUELA TOMÁS HERRERA 5 CHITRÉ ESCUELA JUAN T., DEL BUSTO 6 CHITRÉ ESCUELA SERGIO PÉREZ DELGADO 7 CHITRÉ ESCUELA HIPÓLITO PÉREZ TELLO 8 CHITRÉ C.E.B.G. PRESIDENTE JOHN F., KENNEDY 9 CHITRÉ ESCUELA EVELIO DOLORES CARRIZO 10 CHITRÉ INSTITUTO DE MARINA MERCANTE ISTMEÑA 11 CHITRÉ COLEGIO PSICOPEDAGÓGICO BILINGÜE 12 CHITRÉ COLEGIO SAN JUAN BAUTISTA COLEGIO CENTRO DE FORMACIÓN INTEGRAL BILINGÜE DE AZUERO PAPA 13 CHITRÉ FRANCISCO (SOLO ESTUDIANTES DE I° GRADO) 14 CHITRÉ COLEGIO SANTA RITA BILINGÜE SCHOOL 15 PARITA CEBG. DE PARITA 16 PARITA ESC. PUERTO LIMÓN 17 PARITA ESC. DE PARIS 18 PARITA ESC. POTUGA 19 PARITA ESC. LA CONCEPCIÓN 20 PARITA ESC. PORTOBELILLO 21 PARITA ESC. CABUYA 22 PARITA ESC. LOS HIGOS 23 PARITA ESC. PEDERNAL 24 PARITA ESC. LOS CASTILLOS 25 PARITA ESC. LA VALENCIA 26 PARITA ESC. LLANO DE LA CRUZ MARTES 12, DE MARZO DE 2019 # DISTRITO CENTRO EDUCATIVO CENTRO DE PAGO 1 OCÚ COL. RAFAEL QUINTERO VILLARREAL 2 OCÚ ESC. EL GUARUMAL ESCUELA JOSÉ DOLORES CARRIZO 3 OCÚ ESC. EL HATILLO 4 OCÚ ESC. LA CABUYA 5 OCÚ ESC. LLANO LARGO 6 OCÚ ESC. LOS CARATES 7 OCÚ ESC. QUEBRADA DE AGUA 8 OCÚ ESC. LOS REMEDIOS 9 OCÚ ESC. RINCÓN SANTO 10 OCÚ ESC. LASTENIA DE VILLARREAL 11 OCÚ ESC. GREGORIO GIL 12 OCÚ ESC. LOS JARAMILLOS 13 OCÚ ESC. -
Herrera Reprogramación Pandemia 2020
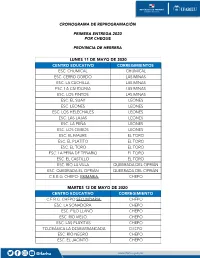
CRONOGRAMA DE REPROGRAMACIÓN PRIMERA ENTREGA 2020 POR CHEQUE PROVINCIA DE HERRERA LUNES 11 DE MAYO DE 2020 CENTRO EDUCATIVO CORREGIMIENTOS ESC. CHUMICAL CHUMICAL ESC. CERRO GORDO LAS MINAS ESC. LA CUCHILLA LAS MINAS ESC. LA CALIDONIA LAS MINAS ESC. LOS PINTOS LAS MINAS ESC. EL SUAY LEONES ESC. LEONES LEONES ESC. LOS HELECHALES LEONES ESC. LAS LAJAS LEONES ESC. LA PEÑA LEONES ESC. LOS CEIBOS LEONES ESC. EL MAURE EL TORO ESC. EL PLATITO EL TORO ESC. EL TORO EL TORO ESC. LA PEÑA DE TEBARIO EL TORO ESC. EL CASTILLO EL TORO ESC. RÍO LA VILLA QUEBRADA DEL CIPRIÁN ESC. QUEBRADA EL CIPRIÁN QUEBRADA DEL CIPRIÁN C.E.B.G. CHEPO- PRIMARIA CHEPO MARTES 12 DE MAYO DE 2020 CENTRO EDUCATIVO CORREGIMIENTO C.E.B.G. CHEPO SECUNDARIA CHEPO ESC. LA SONADORA CHEPO ESC. FILO LLANO CHEPO ESC. RÍO VIEJO CHEPO ESC. LAS PLAYITAS CHEPO TELEBÁSICA LA DESBARRANCADA CHEPO ESC. RÍO NEGRO CHEPO ESC. EL JACINTO CHEPO www.ifarhu.gob.pa ESC. QUEBRADA DEL CÁNTARO QUEBRADA DEL ROSARIO ESC. LOS VIROTALES QUEBRADA DEL ROSARIO ESC. LAS MATAS QUEBRADA DEL ROSARIO ESC. EL NANZAL QUEBRADA DEL ROSARIO ESC. LAS YESCAS QUEBRADA DEL ROSARIO ESC. EL GALLO QUEBRADA DEL ROSARIO ESC. QUEBRADA EL ROSARIO ARRIBA QUEBRADA DEL ROSARIO ESC. QUEBRADA EL ROSARIO ABAJO QUEBRADA DEL ROSARIO ESC. LA HUACA QUEBRADA DEL ROSARIO ESC. EL COPÉ QUEBRADA DEL ROSARIO ESC. LOMA DEL RANCHITO QUEBRADA DEL ROSARIO ESC. EL ALGODÓN QUEBRADA DEL ROSARIO MIERCOLES 13 DE MAYO DE 2020 CENTROS EDUCATIVOS CORREGIMIENTOS ESC. LOS CERRITOS LOS CERRITOS TELEBÁSICA CRUCE DE SANTA CLARA LA PITALOZA ESC. -

Que Crea El Corregimiento De Entradero Del Castillo Segregado Del Corregimiento De Cerro Largo, Distrito De Ocú, Provincia De Herrera
Asamblea Nacional Secretaría General TRÁMITE LEGISLATIVO 2011 ANTEPROYECTO DE LEY: 091 PROYECTO DE LEY: LEY: GACETA OFICIAL: TÍTULO: QUE CREA EL CORREGIMIENTO DE ENTRADERO DEL CASTILLO SEGREGADO DEL CORREGIMIENTO DE CERRO LARGO, DISTRITO DE OCÚ, PROVINCIA DE HERRERA. FECHA DE PRESENTACIÓN: 4 DE OCTUBRE DE 2011. PROPONENTE: HD. JUAN MIGUEL RÍOS. COMISIÓN: ASUNTOS MUNICIPALES. Apartado 0815-01603 Panamá 4 , Panamá Panamá, 3 de octubre de 2011. Honorable Diputado HÉCTOR E. APARICIO DíAZ Presidente de la Asamblea Nacional Respetado Presidente: En ejercicio de la iniciativa legislativa que me otorga el artículo 165 de la ConstitiiCion Política y el artículo 108 del Reglamento Orgánico del Régimen Interno, presento a la consideración de la Asamblea Nacional el Anteproyecto de Ley, Que crea el corregimiento de Entradero del Castillo, segregado del corregimiento de Cerro Largo, distrito de Ocú, provincia de Berrera, el cual merece la siguiente: EXPOSICIÓN DE MOTIVOS El corregimiento de Cerro Largo tiene una superficie de 106.5 kilómetros cuadrados, localizado al sur del distrito de Ocú y hace honor al nombre de una extensa cordillera que se encuentra a lo largo del lugar. Cuenta con un clima tropical húmedo, la precipitación pluvial es de 2000 mm y tiene una temperatura promedio de 28° C y la mínima de 24° C y un promedio de 26°C. Sus límites son: al Norte, con los corregimientos de Los Llanos y Ocú cabera, al Sur, con el distrito de Las Minas (Corrimientos de Toro y Leones), al Este, con el corregimiento de Ocú cabecera, Menchaca y Las Minas y al Oeste, con los corregimientos de Los Llanos y El Tijera. -

Libro Origen Del Nombre De Los Corregimientos
1 2 Orígen del nombre de los corregimientos Magistrados del Tribunal Electoral Erasmo Pinilla C., presidente Eduardo Valdés Escoffery, vicepresidente Heriberto Araúz Sánchez, vocal Magistradas suplentes Lourdes González M. Sharon Sinclaire de Dumanoir Myrtha Varela de Durán Dirección de Comunicación Humberto Castillo M. - Director Daniel Carrasco - Subdirector Dirección Nacional de Oganización Electoral Osman Valdés - Director Santana Díaz - Subdirector Editores Jorge D. Bravo - Tomás Mosquera Diseño y Diagramación Víctor M. Castillo G. Fotografía Tomás Mosquera - Víctor M. Castillo G. Justo Marín Investigación Simón Bolívar Pinto - Direcciones regionales del TE Correctores: Simón Bolívar Pinto - Rodolfo de Gracia Agradecimiento al Sr. Samuel Soane, jefe de Cartografía y al Lcdo. Alonso Ortíz de Zevallos, asesor legal de OE. por la asesoría brindada en esta investigación Impresión: Imprenta del Tribunal Electoral Todos los Derechos Reservados © Diciembre 2014 ÍNDICE Introducción 7/8 Provincia de Chiriquí 58 Distrito de Alanje 58 Provincia de Bocas del Toro 12 Distrito de Barú 61 Distrito de Bocas del Toro 12 Distrito de Boquerón 62 Distrito de Changuinola 13 Distrito de Boquete 65 Distrito de Chiriquí Grande 19 Distrito de Bugaba 69 Distrito de David 75 Provincia de Coclé 24 Distrito de Dolega 78 Distrito de Aguadulce 24 Distrito de Gualaca 81 Distrito de Antón 26 Distrito de Remedios 86 Distrito de La Pintada 31 Distrito de Renacimiento 87 Distrito de Natá 32 Distrio de San Félix 90 Distrito de Olá 35 Distrito de San Lorenzo 91 Distrito -

Cuadro 21. LONGITUD DE LA RED VIAL EN LA REPÚBLICA, SEGÚN PROVINCIA Y COMARCA INDÍGENA, TIPO DE SUPERFICIE Y VÍA: AL 31 DE DICIEMBRE DE 2016 (P)
Cuadro 21. LONGITUD DE LA RED VIAL EN LA REPÚBLICA, SEGÚN PROVINCIA Y COMARCA INDÍGENA, TIPO DE SUPERFICIE Y VÍA: AL 31 DE DICIEMBRE DE 2016 (P) Longitud de la red vial Provincia y comarca indígena, tipo de superficie y vía (en kilómetros) TOTAL 16,366.34 Bocas del Toro 458.23 Asfalto 206.40 División Continental-Chiriquí Grande 35.00 Almirante-Changuinola 21.00 Changuinola-Guabito 13.50 Finca 31-Theobrama 2.70 El Empalme-Charagre 6.70 (Changuinola-Guabito)-Débora-California 6.50 Finca 8-Finca 30 4.20 Finca 30-Finca 31 1.50 Finca 6-Finca 44 4.50 (Changuinola-Almirante)-Ojo de Agua 3.70 Ojo de Agua-Nance Risco 12.00 Finca 4-Finca 3 4.50 Changuinola-Finca 6 1.60 Changuinola-Finca 60 6.00 Finca 60-Finca 61 1.50 Guabito-Las Tablas 18.50 Punta Peña-Almirante 63.00 Tratamiento superficial 63.00 El Empalme-El Silencio 7.50 (El Empalme-Charagre)-Finca 30 0.50 Finca 8-Finca 30 1.80 Bocas del Toro-Boca del Drago 8.00 Bocas del Toro-Big Creek-Punch 2.00 (Punta Peña-Almirante)-Valle Risco 6.50 (Punta Peña-Almirante)-Valle Las Perlas 0.70 (Punta Peña-Almirante)-Valle de Agua 1.80 Las Tablas-Loma del Tigre 6.00 Changuinola-Finca 6 1.30 (Changuinola-Almirante)-La Gloria 1.90 (Changuinola-Almirante)-Junquito 7.50 (Punta Peña-Almirante)-Punta 6.55 (Punta Peña-Almirante)-Valle Sarón 0.25 (Punta Peña-Almirante)-Quebrada Garza 0.20 (Punta Peña-Almirante)-Loma Estrella 0.65 (Punta Peña-Almirante)-Valle Risco-Río Oeste Arriba 1.95 Nance Risco-Charco La Playa 2.60 Nance Risco-Valle Rey 1.30 Cañaza-Higuerones 2.50 Chiriquí Grande-Ballena 0.90 Finca 31-Theobrama 0.60 Revestido 154.23 (El Empalme-Charagre)-Santa Marta-Rómulo 9.50 (El Empalme-Charagre)-Finca 30 1.00 Charagre-Teribe (Quebrada Carbón) 2.50 Cuadro 21. -

Boletín Nº10
ESTADÍSTICA PANAMEÑA SITUACIÓN DEMOGRÁFICA ESTIMACIONES Y PROYECCIONES DE LA POBLACIÓN EN LA REPÚBLICA DE PANAMÁ, POR PROVINCIA, COMARCA INDÍGENA, DISTRITO Y CORREGIMIENTO, SEGÚN SEXO: AÑOS 2000-2015 BOLETÍN N° 10 ÍNDICE Página Metodología 1 Importancia de las estimaciones de población a nivel de corregimiento 5 Análisis de los resultados 7 Estimaciones y Proyecciones de la población en la República por Provincia, Comarca Indígena, Distrito y Corregimiento, según sexo: años 2000-2015 11 Cuadro 1. Estimación de la población total en la República, por provincia, comarca indígena, distrito, corregimiento y sexo: años 2000-2015 13 Cuadro 2. Estimación de la población en la provincia de Bocas del Toro, según distrito, corregimiento y sexo: años 2000-2015 14 Cuadro 3. Estimación de la población en la provincia de Coclé, según distrito, corregimiento y sexo: años 2000-2015 16 Cuadro 4. Estimación de la población en la provincia de Colón, según distrito, corregimiento y sexo: años 2000-2015 20 Cuadro 5. Estimación de la población en la provincia de Chiriquí, según distrito, corregimiento y sexo: años 2000-2015 24 Cuadro 6. Estimación de la población en la provincia de Darién, según distrito, corregimiento, comarca indígena y sexo: años 2000-2015 32 Cuadro 7. Estimación de la población en la provincia de Herrera, según distrito, corregimiento y sexo: años 2000-2015 34 Cuadro 8. Estimación de la población en la provincia de Los Santos, según distrito, corregimiento y sexo: años 2000-2015 38 Cuadro 9. Estimación de la población en la provincia de Panamá, según distrito, corregimiento, comarca indígena y sexo: años 2000-2015 44 Cuadro 10. -

Dirección Provincial De Herrera
HORARIO DE ATENCIÓN: SEGUNDO PAGO 2021 8:00 A.M. - 4:00 P.M. BECASPrimaria, Premedia Y PASE-U y Media DIRECCIÓN PROVINCIAL DE HERRERA LUNES 30 DE AGOSTO DE 2021 DISTRITO CORREGIMIENTO CENTRO DE PAGO CENTRO EDUCATIVO COLEGIO AGUSTINIANO COLEGIO AGUSTINIANO CHITRÉ CHITRÉ PRIMARIA IFARHU COLEGIO SOYUZ MARTES 31 DE AGOSTO DE 2021 DISTRITO CORREGIMIENTO CENTRO DE PAGO CENTRO EDUCATIVO COLEGIO AGUSTINIANO CHITRÉ CHITRÉ COLEGIO AGUSTINIANO PREMEDIA Y MEDIA C.E.F.I.B. DE AZUERO PAPA FRANCISCO C.E.B.G. PRESIDENTE JOHN F. KENNEDY CENT. EDUC. SANTA RITA BILINGUAL SCHOOL COL. SAN JUAN BAUTISTA CENT. ELADIA R. DE COLLADO COL. PSICOPEDAGÓGICO BILINGÜE C.E.B.G. PRESIDENTE JOHN INST. DE MARINA MERCANTE ISTMEÑO - CHITRÉ LA ARENA F. KENNEDY HERRERA INST. NACIONAL DE CULTURA - HERRERA INST. SAN MIGUEL ARCÁNGEL I.P.H.E. CHITRÉ PANASYSTEM ESC. NOCTURNA OFICIAL DE CHITRÉ C.O.I.F. REINTEGRA COL. JOSÉ D. CRESPO ESC. HIPÓLITO PÉREZ TELLO CHITRÉ CHITRÉ ESC. TOMÁS HERRERA ESC. JUAN T. DEL BUSTO ESC. TOMÁS HERRERA COL. PADRE SEGUNDO FAMILIAR CANO ESC. ENEIDA M. DE CASTILLERO COL. PADRE SEGUNDO CHITRÉ CHITRÉ ESC. EVELIO D. CARRIZO FAMILIAR CANO ESC. BOCA DE PARITA ESC. SERGIO PÉREZ DELGADO MIÉRCOLES 1 DE SEPTIEMBRE DE 2021 DISTRITO CORREGIMIENTO CENTRO DE PAGO CENTRO EDUCATIVO LAS MINAS C.E.B.G. LAS MINAS ESC. GLORÍELA BARRÍA LAS MINAS ESC. CERRO GORDO ESC. CHUMICAL ESC. LA CUCHILLA CANCHA MUNICIPAL DE LEONES ESC. LOS HELECHALES LAS MINAS ESC. LA CALIDONIA LAS MINAS ESC. LOS PINTOS ESC. EL SUAY LEONES ESC. LAS LAJAS CHEPO C.E.B.G. -

Hacinamiento En Panamá
Hacinamiento en Panamá Ministerio de Economía y Finanzas Frank De Lima Ministro Omar Castillo Mahesh Khemlani Viceministro de Economía Viceministro de Finanzas Notas aclaratorias En caso de utilizar el material contenido en este informe, agradeceremos citar la fuente o acreditar la autoría al Ministerio de Economía y Finanzas. Signos convencionales que se emplean con mayor frecuencia en la publicación: . Para separar decimales. , Para la separación de millares, millones, etc. .. Dato no aplicable al grupo o categoría. … Información no disponible. - Cantidad nula o cero. 0 Cuando la cantidad es menor a la mitad de la unidad o fracción decimal adoptada 0.0 para la expresión del dato. 0.00 (P) Cifras preliminares o provisionales. (R) Cifras revisadas. (E) Cifras estimadas. n.c.p. No clasificable en otra parte. n.e. No especificado. n.e.p. No especificado en otra partida. n.e.o.c. No especificado en otra categoría. n.e.o.g. No especificado en otro grupo. n.i.o.p. No incluida en otra partida. msnm Metros sobre el nivel del mar B/. Balboa, unidad monetaria del país. Debido al redondeo del computador, la suma o variación puede no coincidir con la cifra impresa Hacinamiento en Panamá Por: Argelis Almanza G. El hacinamiento es interpretado por el vecindad. Se excluyeron las viviendas número de dormitorios con que cuenta la que en el cuestionario no contaban con vivienda, así como por el número de per- información sobre el número de cuartos sonas que ahí suelen exclusivos para dormir pernoctar. y las colectivas que en este caso, no son más El Censo de 2010 re- que hoteles o viviendas gistró 3,405,813 per- temporales. -

116,411 116,828 117,193 117,530 117,826 118,090 118,334 118,551
Cuadro 83. RESUMEN DE LA ESTIMACIÓN Y PROYECCIÓN DE LA POBLACIÓN TOTAL DE LA PROVINCIA DE HERRERA, SEGÚN DISTRITO, CORREGIMIENTO Y SEXO: AÑOS 2010-20 Estimación al 1 de julio Distrito, corregimiento y sexo 2010 2011 2012 2013 2014 2015 2016 2017 2018 2019 2020 TOTAL……………………………………..116,411 116,828 117,193 117,530 117,826 118,090 118,334 118,551 118,736 118,865 118,982 Hombres:………………………………………58,637 58,809 58,948 59,075 59,177 59,269 59,351 59,512 59,655 59,776 59,881 Mujeres………………………………………….57,774 58,019 58,245 58,455 58,649 58,821 58,983 59,039 59,081 59,089 59,101 Chitré….………………………………………………..53,696 54,160 54,602 55,009 55,400 55,794 55,988 56,172 56,344 56,475 56,568 Hombres:……………………………………………………………………25,765 25,967 26,157 26,326 26,497 26,664 26,732 26,841 26,938 27,020 27,102 Mujeres:…………………………………………………………………….27,931 28,193 28,445 28,683 28,903 29,130 29,256 29,331 29,406 29,455 29,466 Chitré (Cabecera)………………………………9,634 9,731 9,826 9,911 9,991 10,079 10,131 10,179 10,223 10,268 10,299 Hombres:……………………………………………………………………4,564 4,604 4,642 4,677 4,706 4,743 4,761 4,784 4,807 4,827 4,848 Mujeres:…………………………………………………………………….5,070 5,127 5,184 5,234 5,285 5,336 5,370 5,395 5,416 5,441 5,451 La Arena………………………………………..8,036 8,098 8,166 8,214 8,275 8,331 8,358 8,377 8,394 8,408 8,413 Hombres:……………………………………………………………………3,872 3,899 3,930 3,949 3,977 4,004 4,017 4,031 4,041 4,053 4,062 Mujeres:…………………………………………………………………….4,164 4,199 4,236 4,265 4,298 4,327 4,341 4,346 4,353 4,355 4,351 Monagrillo………………………………………….13,121 13,232 13,329 13,432 13,518 13,605 -

No. PROVINCIA O COMARCA DISTRITO CORREGIMIENTO
CUADRO No.2 CONTRALORIA GENERAL DE LA REPÚBLICA DIRECCIÓN NACIONAL DE FISCALIZACIÓN GENERAL REPRESENTANTES DE CORREGIMIENTOS DE PANAMÁ QUE GOZAN DE LICENCIA CON SUELDO QUINQUENIO 2019-2024 No. PROVINCIA O COMARCA DISTRITO CORREGIMIENTO REPRESENTANTE CÉDULA LICENCIA CON SUELDO MONTO 1 BOCAS DEL TORO CHANGUINOLA Barriada 4 de abril García Ábrego Ábrego 1-706-988 MINISTERIO DE GOBIERNO 600.00 MINISTERIO DE DESARROLLO 2 BOCAS DEL TORO CHANGUINOLA Finca 30 Antonio Medina 1-24-2301 1,500.00 SOCIAL 3 BOCAS DEL TORO CHANGUINOLA Finca 6 Dudley Owens 1-22-1559 MINISTERIO DE EDUCACION 1,940.30 4 CHIRIQUÍ ALANJE Guarumal Edgar Smith 4-117-52 Aduanas 480.00 5 CHIRIQUÍ ALANJE Palo Grande Efraín Quintero 4-144-454 ANATI 600.00 6 CHIRIQUÍ ALANJE Santo Tomás Karelis Castrejón 4-757-483 PRESIDENCIA DE LA REPUBLICA 2,500.00 7 CHIRIQUÍ BARÚ Limones Aurelio Avilez 4-221-478 AMP 600.00 8 CHIRIQUÍ BOQUERÓN Cordillera Luis Alberto Gómez 8-520-766 MINISTERIO DE EDUCACION 1,929.34 9 CHIRIQUÍ BOQUETE Jaramillo José Manuel Montenegro 4-701-147 MEDUCA 1,852.83 10 CHIRIQUÍ BUGABA Bugaba Edilsa Gómez de Camarena 4-264-258 UNACHI 2,005.60 11 CHIRIQUÍ BUGABA Gómez Ariel Omar Espinosa 4-207-631 PRESIDENCIA DE LA REPUBLICA 600.00 12 CHIRIQUÍ BUGABA La Concepción (cab) Geovani González 4-292-28 MEDUCA 1,940.47 13 CHIRIQUÍ BUGABA San Andrés Edwin Martínez 4-716-614 MEDUCA 1,761.38 14 CHIRIQUÍ BUGABA Santa Marta Roger Martínez 4-261-184 MEDUCA 1,808.67 Osman Daniel Samudio 15 CHIRIQUÍ DAVID Cochea 4-702-375 MINISTERIO DE CULTURA 1,921.10 Guerra 16 CHIRIQUÍ DAVID David Sur -

Enum Instruct Pa1980a.Pdf
INTRODUCCION La finalidad de un Censo de Poblacion y de Vivienda consiste en aten- der ciertas necesidadesde datos estadisticos en un pais dado, necesidades que constituyen los facto res determinantes del contenido del censo. Sin em- bargo, tanto el contenido de estos como las operacionesque se emprenden para obtener los datos deseadosy ponerlos a la disposicion de quienes 10ne- cesitan, exiqen que dichos censos sean servidos par EMPADRONADORES capaces,bien adiestrados y que poseanun alto sentido de honradez, seriedad, espiritu de cooperacion y buena voluntad para trabajar. Usted, como EMPADRONADOR, constituye una unidad en la vasta or- ganizacion censal que tiene ramificaciones en todo el territorio nacional; par 10 tanto es de suma importancia que estudie con detenimiento, antes de ini- ciar su labor, este Manual que pone a su disposicion una serie de normas, ex- plicaciones, recomendaciones y elementos basicos para la interpretacion del material cartografico que Ie permitiran realizar una eficiente labor en los Censosde Poblacion y Vivienda que se levantaran en el pais el 11 de mayo de 1980. CONTENIDO Pagina numero ASPECTOSGENERALES 1. Que es un Censo de Poblacion y un Censode Vivienda . 1 2. Par que se levanta un Censo .. 1 3. Finalidad basica de 105Censos de Poblacion y Vivienda . 1 4. A quienes empadronarael Censo. 2 5. Fecha y duracion del empadronamiento . 3 6. Organizacion del Empadronamiento . 3 7. Material que usarael Empadronador . 4 DE LOS DATOS 1. Obligation de suministrar los datos. 5 2. Confidencialidad de los datos. 6 OBLIGACIONES RESPECTOA LOS CENSOS Algunos art(culos del Decreto-Ley No.7 de 25 de Febrero de 1960. -

Línea-Base-Río-Parita-130.Pdf
Línea Base: diagnóstico biofísico, socioeconómico y potencial energético de la Cuenca del Río Parita 2012 Consultor: Ingeniero Ivanor Ruiz de León Página 1 Línea Base: diagnóstico biofísico, socioeconómico y potencial energético de la Cuenca del Río Parita 2012 Preparado por Ingeniero Ivanor Ruiz De León Equipo responsable Coordinador Ingeniero Ivanor Ruiz De León Biólogo Jorge Mendieta Ingeniero Forestal Octavio Carrasquilla Ingeniero Ambiental Euclides Domínguez Ingeniero Geólogo Alberto Einsten Ruiz Ingeniero Agrónomo Blas Felipe Morán Sociólogo Julio Ernesto Moreno Geógrafo Carlos Calderón Diciembre de 2012 Consultor: Ingeniero Ivanor Ruiz de León Página 2 Línea Base: diagnóstico biofísico, socioeconómico y potencial energético de la Cuenca del Río Parita 2012 Índice Pag. Resumen Ejecutivo 14 Introducción 22 Objetivos 23 Metodología 23 Trabajo en campo 25 Análisis de información obtenida 26 Preparación del informe 27 Metodología utilizada para la elaboración de los mapas para la cuenca hidrográfica del río Parita 28 1. Aspectos biológicos 33 1.1. Flora 33 1.1.1. Cobertura boscosa 34 1.1.2. Nivel de deforestación de la cuenca 39 1.1.3. Tipos de vegetación y especies dominantes 37 1.1.3.1. Manglar 38 1.1.3.2. Bosque intervenido 39 1.1.3.3. Bosque maduro 42 1.1.3.4. Vegetación baja inundable 43 1.1.3.5. Rastrojo 45 1.1.3.6. Otros usos 46 1.1.4. Ecosistemas frágiles 46 1.1.5. Áreas protegidas 47 1.1.5.1. El Parque Nacional Sarigua 47 1.1.5.2. Reserva Forestal Cerro Camarón y Pedregoso 48 1.1.5.3. Zona de Protección Territorial, Urbana y Ambiental sector costero de Chitré 48 1.1.6.