Ascidian” Mitochondrial Genetic Code in Tunicates [Version 1; Peer Review: 2 Approved] Julien Pichon 1,2, Nicholas M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Pyura Doppelgangera to Support Regional Response Decisions
REPORT NO. 2480 BACKGROUND INFORMATION ON THE SEA SQUIRT PYURA DOPPELGANGERA TO SUPPORT REGIONAL RESPONSE DECISIONS CAWTHRON INSTITUTE | REPORT NO. 2480 JUNE 2014 BACKGROUND INFORMATION ON THE SEA SQUIRT PYURA DOPPELGANGERA TO SUPPORT REGIONAL RESPONSE DECISIONS LAUREN FLETCHER Prepared for Marlborough District Council CAWTHRON INSTITUTE 98 Halifax Street East, Nelson 7010 | Private Bag 2, Nelson 7042 | New Zealand Ph. +64 3 548 2319 | Fax. +64 3 546 9464 www.cawthron.org.nz REVIEWED BY: APPROVED FOR RELEASE BY: Javier Atalah Chris Cornelisen ISSUE DATE: 3 June 2014 RECOMMENDED CITATION: Fletcher LM 2014. Background information on the sea squirt, Pyura doppelgangera to support regional response decisions. Prepared for Marlborough District Council. Cawthron Report No. 2480. 30 p. © COPYRIGHT: Cawthron Institute. This publication may be reproduced in whole or in part without further permission of the Cawthron Institute, provided that the author and Cawthron Institute are properly acknowledged. CAWTHRON INSTITUTE | REPORT NO. 2480 JUNE 2014 EXECUTIVE SUMMARY The non-indigenous solitary sea squirt, Pyura doppelgangera (herein Pyura), was first detected in New Zealand in 2007 after a large population was found in the very north of the North Island. A delimitation survey by the Ministry for Primary Industries (MPI) during October 2009 found established populations at 21 locations within the region. It is not known how long Pyura has been present in New Zealand, although it is not believed to be a recent introduction. Pyura is an aggressive interspecific competitor for primary space. As such, this species may negatively impact native green-lipped mussel beds present, with associated impacts to key social and cultural values. -
Appendicularia of CTAW
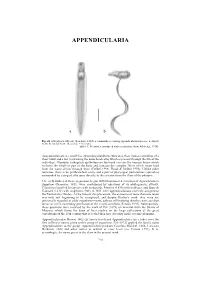
APPENDICULARIA APPENDICULARIA a b Fig. 26. Oikopleura albicans (Leuckart, 1854), a commonly occurring appendicularian species: a, dorsal view; b, lateral view. (Scale bar = 0.1 mm). [after T. Prentiss, reproduced with permission, from Alldredge 1976] Appendicularians are small free swimming planktonic tunicates, their bodies consisting of a short trunk and a tail (containing the notochord cells) which is present through the life of the individual. Glandular (oikoplast) epithelium on the trunk secretes the mucous house which encloses the whole or part of the body and contains the complex filters which strain food from the water driven through them (Deibel 1998; Flood & Deibel 1998). Unlike other tunicates, there is no peribranchial cavity and a pair of pharyngeal perforations (spiracles) surrounded by a ring of cilia open directly to the exterior from the floor of the pharynx. The early studies of these organisms, begun with Chamisso's description of Appendicularia flagellum Chamisso, 1821, were confounded by questions of its phylogenetic affinity. Chamisso classified his species with medusoids, Mertens (1830) with molluscs, and Quoy & Gaimard (1833) with zoophytes. Only in 1851 were appendicularians correctly assigned to the Tunicata by Huxley. At the time of this placement, the existence of more than one taxon was only just beginning to be recognised, and despite Huxley's work, they were not universally regarded as adult organisms—some authors still insisting that they were ascidian larvae or a free swimming generation of the sessile ascidians (Fenaux 1993). Subsequently, these questions were resolved by the work of Fol (1872) on material from the Straits of Messina, which forms the basis of later studies on the large collections of the great expeditions of the 19th century that revealed their true diversity in the oceanic plankton. -

Study of Dental Fluorosis in Subjects Related to a Phosphatic Fertilizer
Indian Journal of Geo Marine Sciences Vol. 46 (07), July 2017, pp. 1371-1380 Diversity and abundance of epipelagic larvaceans and calanoid copepods in the eastern equatorial Indian Ocean during the spring inter-monsoon Kaizhi Li, Jianqiang Yin*, Yehui Tan, Liangmin Huang & Gang Li Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China *[E-mail: [email protected]] Received 20 August 2015; revised 29 September 2015 This study investigated the species composition, distribution and abundance of larvaceans and calanoid copepods in the eastern equatorial Indian Ocean. In total, 25 species of larvaceans and 69 species of calanoid copepods were identified in the study area. Although the average diversity and evenness indexes of larvaceans were lower than those of calanoid copepods, the abundance of larvaceans was higher than that of calanoids with means of 40.1±14.9 ind m-3 and 28.4±9.1 ind m-3, respectively. Larvacean community was numerically dominated by Oikopleura fusiformis, Oikopleura longicauda, Oikopleura cophocerca, Fritillaria formica and Fritillaria pellucida, accounting for 83% of total larvacean abundance. The calanoid community was dominated by the following five species, represented 61% of calanoid copepods: Clausocalanus furcatus, Clausocalanus farrani, Acartia negligens, Acrocalanus longicornis as well as the copepodite stage of Euchaeta spp. This study highlights that the importance of larvaceans in the eastern equatorial Indian Ocean. [Keywords: Appendicularians, Calanoids, Water mass, Monsoon, Indian Ocean] Introduction It has been clear that small organisms of marine Clausocalanus) and the cyclopoid genera (such as zooplankton have historically been under-sampled by Oithona, Oncaea and Corycaeus1). -

Science Journals
SCIENCE ADVANCES | RESEARCH ARTICLE MARINE ECOLOGY Copyright © 2017 The Authors, some From the surface to the seafloor: How giant larvaceans rights reserved; exclusive licensee transport microplastics into the deep sea American Association for the Advancement † † of Science. No claim to Kakani Katija,* C. Anela Choy,* Rob E. Sherlock, Alana D. Sherman, Bruce H. Robison original U.S. Government Works. Distributed Plastic waste is a pervasive feature of marine environments, yet little is empirically known about the biological under a Creative and physical processes that transport plastics through marine ecosystems. To address this need, we conducted Commons Attribution in situ feeding studies of microplastic particles (10 to 600 mm in diameter) with the giant larvacean Bathochordaeus NonCommercial stygius. Larvaceans are abundant components of global zooplankton assemblages, regularly build mucus “houses” License 4.0 (CC BY-NC). to filter particulate matter from the surrounding water, and later abandon these structures when clogged. By conducting in situ feeding experiments with remotely operated vehicles, we show that giant larvaceans are able to filter a range of microplastic particles from the water column, ingest, and then package microplastics into their fecal pellets. Microplastics also readily affix to their houses, which have been shown to sink quickly to the seafloor and deliver pulses of carbon to benthic ecosystems. Thus, giant larvaceans can contribute to the vertical flux of microplastics through the rapid sinking of fecal pellets and discarded houses. Larvaceans, and potentially other abundant pelagic filter feeders, may thus comprise a novel biological transport mechanism delivering microplastics from surface waters, through the water column, and to the seafloor. -

Mitochondrial Genetic Code in Tunicates
bioRxiv preprint doi: https://doi.org/10.1101/865725; this version posted December 5, 2019. The copyright holder has placed this preprint (which was not certified by peer review) in the Public Domain. It is no longer restricted by copyright. Anyone can legally share, reuse, remix, or adapt this material for any purpose without crediting the original authors. Widespread use of the \ascidian" mitochondrial genetic code in tunicates Julien Pichon1,2, Nicholas M. Luscombe1,3,4, Charles Plessy1,* December 5, 2019 1. Okinawa Institute of Science and Technology Graduate University 1919-1 Tancha, Onna-son, Kunigami-gun Okinawa, 904-0495, Japan 2. Uni- versit´ede Paris 3. The Francis Crick Institute, 1 Midland Road, Lon- don, NW1 1AT, UK 4. UCL Genetics Institute, University College Lon- don, Gower Street, London, WC1E 6BT, UK *. Author for correspondence: [email protected] Abstract Background: Ascidians, a tunicate class, use a mitochondrial ge- netic code that is distinct from vertebrates and other invertebrates. Though it has been used to translate the coding sequences from other tunicate species on a case-by-case basis, it is has not been investi- gated whether this can be done systematically. This is an impor- tant because a) some tunicate mitochondrial sequences are currently translated with the invertebrate code by repositories such as NCBI's GenBank, and b) uncertainties about the genetic code to use can com- plicate or introduce errors in phylogenetic studies based on translated mitochondrial protein sequences. Methods: We collected publicly available nucleotide sequences for non-ascidian tunicates including appendicularians such as Oikopleura dioica, translated them using the ascidian mitochondrial code, and built multiple sequence alignments covering all tunicate classes. -

Tunicate Mitogenomics and Phylogenetics: Peculiarities of the Herdmania Momus Mitochondrial Genome and Support for the New Chordate Phylogeny
Tunicate mitogenomics and phylogenetics: peculiarities of the Herdmania momus mitochondrial genome and support for the new chordate phylogeny. Tiratha Raj Singh, Georgia Tsagkogeorga, Frédéric Delsuc, Samuel Blanquart, Noa Shenkar, Yossi Loya, Emmanuel Douzery, Dorothée Huchon To cite this version: Tiratha Raj Singh, Georgia Tsagkogeorga, Frédéric Delsuc, Samuel Blanquart, Noa Shenkar, et al.. Tu- nicate mitogenomics and phylogenetics: peculiarities of the Herdmania momus mitochondrial genome and support for the new chordate phylogeny.. BMC Genomics, BioMed Central, 2009, 10, pp.534. 10.1186/1471-2164-10-534. halsde-00438100 HAL Id: halsde-00438100 https://hal.archives-ouvertes.fr/halsde-00438100 Submitted on 2 Dec 2009 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. BMC Genomics BioMed Central Research article Open Access Tunicate mitogenomics and phylogenetics: peculiarities of the Herdmania momus mitochondrial genome and support for the new chordate phylogeny Tiratha Raj Singh†1, Georgia Tsagkogeorga†2, Frédéric Delsuc2, Samuel Blanquart3, Noa -

Title GENERAL CONSIDERATION on JAPANESE APPENDICULARIAN
GENERAL CONSIDERATION ON JAPANESE Title APPENDICULARIAN FAUNA Author(s) Tokioka, Takasi PUBLICATIONS OF THE SETO MARINE BIOLOGICAL Citation LABORATORY (1955), 4(2-3): 251-261 Issue Date 1955-05-30 URL http://hdl.handle.net/2433/174523 Right Type Departmental Bulletin Paper Textversion publisher Kyoto University GENERAL CONSIDERATION ON JAPANESE APPENDICULARIAN FAUNA') T AKASI TOKIOKA Seto Marine Biological Laboratory, Sirahama With 6 Text-figures I. Species Occlilrring ilil Japanese Waters and the Neighbolilring Seas. So far as I am aware, the following 34 species of appendicularians are found in reports dealing with this animal group occurring in Japanese waters and the neigh bouring seas. Oikopleuridae Oikopleura (Coecaria) longicauda VoGT (A, T,, T 2 , T 3 ) Oikopleura (Coecaria) fusiformis FoL (A, T,, T 2 , Ts) Oikopleura (Coecaria) intermedia LOHMANN ( =Oik. microstoma AIDA) (A, T,, T 2 ) *Oikopleura (Coecaria) cornutogastra AIDA (A, T 2 , T 3) Oikopleura (Coecaria) graciloides LOHMANN & BuCKMANN (T2 ) Oikopleura (Coecaria) gracilis LOHMANN (T3 ) Oikopleura ( Vexillaria) dioica FoL (A, T,, T 2 , T 3 ) Oikopleura (Vexillaria) rufescens FoL (A, T,, T 2 , T 3) Oikopleura ( Vexillaria) parva LoHMANN (T3 ) Oikopleura (Vexillaria) cophocerca GEGENBAUR (T1 , T 2 , T 3) Oikopleura ( Vexillaria) albicans (LEUCKART) (T,) Oikopleura ( Vexillaria) labradoriensis LoHMANN (T,, T 3 ) *Oikopleura ( Vexillaria) chamissonis MERTENS (MERTENS 1831) Megalocercus huxleyi (RITTER) (=Oik. megastoma AIDA) (A, T,, T 2 , T 3 ) Stegosoma magnum (LANGERHANS) (A, T 1 , T 2 , T 3 ) *Chunopleura microgaster LOHMANN (T,) Pelagopleura verticalis (LoHMANN) (T2 ) *Pelagopleura sp. (T,, T 3 ) Fritillaridae Fritillaria (Acrocercus) haplostoma FoL (A, T,, T 2 , Ts) ----------------------------------·------- 1) Contributions from the Seto Marine Biological Laboratory, No. -

Can Novel Genetic Analyses Help to Identify Lowdispersal Marine
Can novel genetic analyses help to identify low-dispersal marine invasive species? Peter R. Teske1,2, Jonathan Sandoval-Castillo1, Jonathan M. Waters3 & Luciano B. Beheregaray1 1Molecular Ecology Laboratory, School of Biological Sciences, Flinders University, Adelaide, South Australia 5001, Australia 2Department of Zoology, University of Johannesburg, Auckland Park, 2006, Johannesburg, South Africa 3Department of Zoology, University of Otago, PO Box 56, Dunedin, New Zealand Keywords Abstract Ascidian, biological invasion, coalescent theory, founder effect, genetic bottleneck, Genetic methods can be a powerful tool to resolve the native versus introduced microsatellites, sea squirt. status of populations whose taxonomy and biogeography are poorly understood. The genetic study of introduced species is presently dominated by analyses that Correspondence identify signatures of recent colonization by means of summary statistics. Unfor- Luciano B. Beheregaray tunately, such approaches cannot be used in low-dispersal species, in which Molecular Ecology Laboratory, School of recently established populations originating from elsewhere in the species’ native Biological Sciences, Flinders University, range also experience periods of low population size because they are founded by Adelaide, SA 5001, Australia. Tel: +61 8 8201 5243; Fax: +61 8 8201 3015; few individuals. We tested whether coalescent-based molecular analyses that pro- E-mail: Luciano.beheregaray@flinders.edu.au vide detailed information about demographic history supported the hypothesis that a sea squirt whose distribution is centered on Tasmania was recently intro- Funding Information duced to mainland Australia and New Zealand through human activities. Meth- This study was funded by the Australian ods comparing trends in population size (Bayesian Skyline Plots and Research Council (DP110101275 to Approximate Bayesian Computation) were no more informative than summary Beheregaray, Moller€ & Waters). -

Phylum: Chordata
PHYLUM: CHORDATA Authors Shirley Parker-Nance1 and Lara Atkinson2 Citation Parker-Nance S. and Atkinson LJ. 2018. Phylum Chordata In: Atkinson LJ and Sink KJ (eds) Field Guide to the Ofshore Marine Invertebrates of South Africa, Malachite Marketing and Media, Pretoria, pp. 477-490. 1 South African Environmental Observation Network, Elwandle Node, Port Elizabeth 2 South African Environmental Observation Network, Egagasini Node, Cape Town 477 Phylum: CHORDATA Subphylum: Tunicata Sea squirts and salps Urochordates, commonly known as tunicates Class Thaliacea (Salps) or sea squirts, are a subphylum of the Chordata, In contrast with ascidians, salps are free-swimming which includes all animals with dorsal, hollow in the water column. These organisms also ilter nerve cords and notochords (including humans). microscopic particles using a pharyngeal mucous At some stage in their life, all chordates have slits net. They move using jet propulsion and often at the beginning of the digestive tract (pharyngeal form long chains by budding of new individuals or slits), a dorsal nerve cord, a notochord and a post- blastozooids (asexual reproduction). These colonies, anal tail. The adult form of Urochordates does not or an aggregation of zooids, will remain together have a notochord, nerve cord or tail and are sessile, while continuing feeding, swimming, reproducing ilter-feeding marine animals. They occur as either and growing. Salps can range in size from 15-190 mm solitary or colonial organisms that ilter plankton. in length and are often colourless. These organisms Seawater is drawn into the body through a branchial can be found in both warm and cold oceans, with a siphon, into a branchial sac where food particles total of 52 known species that include South Africa are removed and collected by a thin layer of mucus within their broad distribution. -

1 Contemporary Climate Change Hinders Hybrid Performance of Ecologically Dominant
1 Contemporary climate change hinders hybrid performance of ecologically dominant 2 marine invertebrates 3 4 Jamie Hudson1*, Christopher D McQuaid2, Marc Rius1,3 5 6 1 School of Ocean and Earth Science, National Oceanography Centre Southampton, 7 University of Southampton, European Way, Southampton, SO14 3ZH, United Kingdom 8 2 Coastal Research Group, Department of Zoology and Entomology, Rhodes University, G3, 9 South Africa 10 3 Department of Zoology, Centre for Ecological Genomics and Wildlife Conservation, 11 University of Johannesburg, Auckland Park, South Africa 12 * Corresponding email: [email protected] 13 14 Running title: Climate change and hybrid performance 15 Acknowledgements 16 We would like to thank Jaqui Trassierra, Aldwyn Ndhlovu, and Cristian Monaco for their 17 assistance in sample collection. We acknowledge Carlota Fernández Muñiz for her 18 invaluable assistance in the culturing of algae for the experiments, Dr Ivan Haigh for 19 assistance collecting and analysing seawater temperature data, and Prof Dustin Marshall for 20 his help in methodological aspects. Funding for JH’s stay at Rhodes University was provided 21 for by the Research Training and Support Grant from the Southampton Marine and 22 Maritime Institution. This work is based upon research supported by the South African 23 Research Chairs Initiative of the Department of Science and Technology and the National 24 Research Foundation. 1 25 26 Abstract 27 Human activities alter patterns of biodiversity, particularly through species extinctions and 28 range contractions. Two of these activities are human mediated transfer of species and 29 contemporary climate change, and both allow previously isolated genotypes to come into 30 contact and hybridise, potentially altering speciation rates. -

Mitochondrial Genetic Code in Tunicates[Version
F1000Research 2020, 8:2072 Last updated: 23 SEP 2021 RESEARCH ARTICLE Widespread use of the “ascidian” mitochondrial genetic code in tunicates [version 2; peer review: 2 approved] Julien Pichon 1,2, Nicholas M. Luscombe 1,3,4, Charles Plessy 1 1Genomics and Regulatory Systems Unit, Okinawa Institute of Science and Technology Graduate University, Onna-son, Okinawa, 904-0495, Japan 2Université de Paris, Paris, France 3The Francis Crick Institute, London, NW1 1AT, UK 4Genetics Institute, University College London, London, WC1E 6BT, UK v2 First published: 10 Dec 2019, 8:2072 Open Peer Review https://doi.org/10.12688/f1000research.21551.1 Latest published: 14 Apr 2020, 8:2072 https://doi.org/10.12688/f1000research.21551.2 Reviewer Status Invited Reviewers Abstract Background: Ascidians, a tunicate class, use a mitochondrial genetic 1 2 code that is distinct from vertebrates and other invertebrates. Though it has been used to translate the coding sequences from other version 2 tunicate species on a case-by-case basis, it is has not been (revision) investigated whether this can be done systematically. This is an 14 Apr 2020 important because a) some tunicate mitochondrial sequences are currently translated with the invertebrate code by repositories such as version 1 NCBI GenBank, and b) uncertainties about the genetic code to use can 10 Dec 2019 report report complicate or introduce errors in phylogenetic studies based on translated mitochondrial protein sequences. 1. Patrick Lemaire , University of Methods: We collected publicly available nucleotide sequences for non-ascidian tunicates including appendicularians such as Oikopleura Montpellier, Montpellier, France dioica, translated them using the ascidian mitochondrial code, and built multiple sequence alignments covering all tunicate classes. -

(Gulf Watch Alaska) Final Report the Seward Line: Marine Ecosystem
Exxon Valdez Oil Spill Long-Term Monitoring Program (Gulf Watch Alaska) Final Report The Seward Line: Marine Ecosystem monitoring in the Northern Gulf of Alaska Exxon Valdez Oil Spill Trustee Council Project 16120114-J Final Report Russell R Hopcroft Seth Danielson Institute of Marine Science University of Alaska Fairbanks 905 N. Koyukuk Dr. Fairbanks, AK 99775-7220 Suzanne Strom Shannon Point Marine Center Western Washington University 1900 Shannon Point Road, Anacortes, WA 98221 Kathy Kuletz U.S. Fish and Wildlife Service 1011 East Tudor Road Anchorage, AK 99503 July 2018 The Exxon Valdez Oil Spill Trustee Council administers all programs and activities free from discrimination based on race, color, national origin, age, sex, religion, marital status, pregnancy, parenthood, or disability. The Council administers all programs and activities in compliance with Title VI of the Civil Rights Act of 1964, Section 504 of the Rehabilitation Act of 1973, Title II of the Americans with Disabilities Action of 1990, the Age Discrimination Act of 1975, and Title IX of the Education Amendments of 1972. If you believe you have been discriminated against in any program, activity, or facility, or if you desire further information, please write to: EVOS Trustee Council, 4230 University Dr., Ste. 220, Anchorage, Alaska 99508-4650, or [email protected], or O.E.O., U.S. Department of the Interior, Washington, D.C. 20240. Exxon Valdez Oil Spill Long-Term Monitoring Program (Gulf Watch Alaska) Final Report The Seward Line: Marine Ecosystem monitoring in the Northern Gulf of Alaska Exxon Valdez Oil Spill Trustee Council Project 16120114-J Final Report Russell R Hopcroft Seth L.