Naive Time Series Forecasting Methods
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Progress in the R Ecosystem for Representing and Handling Spatial Data
Journal of Geographical Systems https://doi.org/10.1007/s10109-020-00336-0 ORIGINAL ARTICLE Progress in the R ecosystem for representing and handling spatial data Roger S. Bivand1 Received: 9 October 2019 / Accepted: 8 September 2020 © The Author(s) 2020 Abstract Twenty years have passed since Bivand and Gebhardt (J Geogr Syst 2(3):307–317, 2000. https ://doi.org/10.1007/PL000 11460 ) indicated that there was a good match between the then nascent open-source R programming language and environment and the needs of researchers analysing spatial data. Recalling the development of classes for spatial data presented in book form in Bivand et al. (Applied spatial data analysis with R. Springer, New York, 2008, Applied spatial data analysis with R, 2nd edn. Springer, New York, 2013), it is important to present the progress now occurring in representation of spatial data, and possible consequences for spatial data handling and the statistical analysis of spatial data. Beyond this, it is imperative to discuss the relationships between R-spatial software and the larger open-source geospatial software community on whose work R packages crucially depend. Keywords Spatial data analysis · Open-source software · R programming language JEL Classifcation C00 · C88 · R15 1 Introduction While Bivand and Gebhardt (2000) did provide an introduction to R as a statistical programming language and to why one might choose to use a scripted language like R (or Python), this article is both retrospective and prospective. It is possible that those approaching the choice of tools for spatial analysis and for handling spatial data will fnd the following less than inviting; in that case, perusal of early chapters of Lovelace et al. -

Sweave User Manual
Sweave User Manual Friedrich Leisch and R-core June 30, 2017 1 Introduction Sweave provides a flexible framework for mixing text and R code for automatic document generation. A single source file contains both documentation text and R code, which are then woven into a final document containing • the documentation text together with • the R code and/or • the output of the code (text, graphs) This allows the re-generation of a report if the input data change and documents the code to reproduce the analysis in the same file that contains the report. The R code of the complete 1 analysis is embedded into a LATEX document using the noweb syntax (Ramsey, 1998) which is usually used for literate programming Knuth(1984). Hence, the full power of LATEX (for high- quality typesetting) and R (for data analysis) can be used simultaneously. See Leisch(2002) and references therein for more general thoughts on dynamic report generation and pointers to other systems. Sweave uses a modular concept using different drivers for the actual translations. Obviously different drivers are needed for different text markup languages (LATEX, HTML, . ). Several packages on CRAN provide support for other word processing systems (see Appendix A). 2 Noweb files noweb (Ramsey, 1998) is a simple literate-programming tool which allows combining program source code and the corresponding documentation into a single file. A noweb file is a simple text file which consists of a sequence of code and documentation segments, called chunks: Documentation chunks start with a line that has an at sign (@) as first character, followed by a space or newline character. -

ESS — Emacs Speaks Statistics
ESS — Emacs Speaks Statistics ESS version 5.14 The ESS Developers (A.J. Rossini, R.M. Heiberger, K. Hornik, M. Maechler, R.A. Sparapani, S.J. Eglen, S.P. Luque and H. Redestig) Current Documentation by The ESS Developers Copyright c 2002–2010 The ESS Developers Copyright c 1996–2001 A.J. Rossini Original Documentation by David M. Smith Copyright c 1992–1995 David M. Smith Permission is granted to make and distribute verbatim copies of this manual provided the copyright notice and this permission notice are preserved on all copies. Permission is granted to copy and distribute modified versions of this manual under the conditions for verbatim copying, provided that the entire resulting derived work is distributed under the terms of a permission notice identical to this one. Chapter 1: Introduction to ESS 1 1 Introduction to ESS The S family (S, Splus and R) and SAS statistical analysis packages provide sophisticated statistical and graphical routines for manipulating data. Emacs Speaks Statistics (ESS) is based on the merger of two pre-cursors, S-mode and SAS-mode, which provided support for the S family and SAS respectively. Later on, Stata-mode was also incorporated. ESS provides a common, generic, and useful interface, through emacs, to many statistical packages. It currently supports the S family, SAS, BUGS/JAGS, Stata and XLisp-Stat with the level of support roughly in that order. A bit of notation before we begin. emacs refers to both GNU Emacs by the Free Software Foundation, as well as XEmacs by the XEmacs Project. The emacs major mode ESS[language], where language can take values such as S, SAS, or XLS. -

Conference Brochure S P E a K E R S
04.07.2017 – 07.07.2017 www.user2017.brussels @useR_Brussels #user2017 CONFERENCE BROCHURE S P E A K E R S 2 As a student my use of R for data analyses was frowned upon - the suggestion was to stick with the existing software. P Access to R related literature was difficult. To admire R the very first R books one had to travel far, to the few enlightened universities. As a researcher I needed the help E of a secret floppy disk to dual boot into a GNU/Linux system with R, in order to escape from unnamed products. As a F starter in industry, people from unnamed companies made clear to my management that setting up R events for customers was A giving a ‘wrong signal to the market’. C Look how ‘wrong’ we have been... Fifteen years later R is shining brightly in the data science field. Students are E learning R before anything else and if statistics research is published without accompanying R package, it is considered a bad sign. Publishers have hundreds of books on display and major software houses are in a constant race against time to further integrate R into their product offerings. And now, more than 1000 people from all over the world have come to Brussels to discover the state of R at this conference. R has come a great way and the community that formed around the R project is and has always been critical to its success. The community has evolved in many ways and is active in many places throughout the year, but of all things the official useR conference still is the major community event of the year. -

August 12-14, Dortmund, Germany
AASC Austrian Association for Statistical Computing 2008 Program August 12-14, Dortmund, Germany useR! 2008, Dortmund, Germany 1 Contents Greetings and Miscellaneous 2 Maps 5 Social Program 8 Program 9 Tutorials 9 Schedule 10 List of Talks 14 2 useR! 2008, Dortmund, Germany Dear useRs, the following pages provide you with some useful information about useR! 2008, the R user con- ference, taking place at the Fakultät Statistik, Technische Universität Dortmund, Germany from 2008-08-12 to 2008-08-14. Pre-conference tutorials will take place on August 11. The confer- ence is organized by the Fakultät Statistik, Technische Universität Dortmund and the Austrian Association for Statistical Computing (AASC). Apart from challenging and likewise exciting scientific contributions we hope to offer you an attractive conference site and a pleasant social program. With best regards from the organizing team: Uwe Ligges (conference), Achim Zeileis (program), Claus Weihs, Gerd Kopp (local organiza- tion), Friedrich Leisch, and Torsten Hothorn Address / Contact Address: Uwe Ligges Fakultät Statistik Technische Universität Dortmund 44221 Dortmund Germany Phone: +49 231 755 4353 Fax: +49 231 755 4387 e-mail: [email protected] URL: http://www.R-Project.org/useR-2008/ Program Committee Micah Altman, Roger Bivand, Peter Dalgaard, Jan de Leeuw, Ramón Díaz-Uriarte, Spencer Graves, Leonhard Held, Torsten Hothorn, François Husson, Christian Kleiber, Friedrich Leisch, Andy Liaw, Martin Mächler, Kate Mullen, Ei-ji Nakama, Thomas Petzoldt, Martin Theus, and Heather Turner Conference Location Technische Universität Dortmund Campus Nord Mathematikgebäude / Audimax Vogelpothsweg 87 44227 Dortmund Conference Office Opening hours: Monday, August 11: 08:30–19:30 Tuesday, August 12: 08:30–18:30 Wednesday, August 13: 08:30–18:30 Thursday, August 14: 08:30–15:30 useR! 2008, Dortmund, Germany 3 Public Transport The conference site at the university campus and the city hall are best to be reached by public transport. -

Species' Traits and Phylogenetic
Northern Michigan University NMU Commons All NMU Master's Theses Student Works 8-2018 CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES Katie Nehiba [email protected] Follow this and additional works at: https://commons.nmu.edu/theses Part of the Ecology and Evolutionary Biology Commons Recommended Citation Nehiba, Katie, "CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES" (2018). All NMU Master's Theses. 557. https://commons.nmu.edu/theses/557 This Open Access is brought to you for free and open access by the Student Works at NMU Commons. It has been accepted for inclusion in All NMU Master's Theses by an authorized administrator of NMU Commons. For more information, please contact [email protected],[email protected]. CLIMATE DRIVEN RANGE SHIFTS OF NORTH AMERICAN SMALL MAMMALS: SPECIES’ TRAITS AND PHYLOGENETIC INFLUENCES By Katie R. Nehiba THESIS Submitted to Northern Michigan University In partial fulfillment of the requirements For the degree of MASTER OF SCIENCE Office of Graduate Education and Research July 2018 ABSTRACT By Katie R. Nehiba Current anthropogenically-driven climate change is accelerating at an unprecedented rate. In response, species’ ranges may shift, tracking optimal climatic conditions. Species-specific differences may produce predictable differences in the extent of range shifts. I evaluated if patterns of predicted responses to climate change were strongly related to species’ taxonomic identities and/or ecological characteristics of species’ niches, elevation and precipitation. I evaluated differences in predicted range shifts in well-sampled small mammals that are restricted to North America: kangaroo rats, voles, chipmunks, and ground squirrels. -

Reproducible Research.. Using Sweave, Knitr and Pandoc
Reproducible Research.. Using Sweave, Knitr and Pandoc Aedín Culhane [email protected] Nov 20th 2012 My R Course Website http://bcb.dfci.harvard.edu/~aedin/ My HSPH homepage http://www.hsph.harvard.edu/research/aedin-culhane/ When issues of reproducibility arise • ``Remember that microarray analysis you did six months ago? We ran a few more arrays. Can you add them to the project and repeat the same analysis?'' • ``The statistical analyst who looked at the data I generated previously is no longer available. Can you get someone else to analyze my new data set using the same methods (and thus producing a report I can expect to understand)?'' • ``Please write/edit the methods sections for the abstract/paper/grant proposal I am submitting based on the analysis you did several months ago.'' From Keith Baggerly • Selected articles published in Nature Genetics between January 2005 and December 2006 that had used profiling with microarrays • Of the 56 items retrieved electronically, 20 articles were considered potentially eligible for the project • The four teams were from – University of Alabama at Birmingham (UAB) – Stanford/Dana-Farber (SD) – London (L) and Ioannina/Trento (IT) • Each team was comprised of 3-6 scientists who worked together to evaluate each article. Results • Result could be reproduced n=2 • Reproduced with discrepancy n=6 • Could not be reproduced n=10 – No data n=4 (no data n=2, subset n=1, no reporter data n=1) – Confusion over matching of data to analysis (n=2) – Specialized software required and not available (n=1)m – Raw data available but could not be processed n=2 Reproducibility of Analysis Ioannidis JP, Allison DB, Ball CA, Coulibaly I, Cui X, Culhane AC, et al, (2009) Repeatability of published microarray gene expression Analyses. -
Dynamic Documents with R and Knitr, by Yihui Xie 116 Tugboat, Volume 35 (2014), No
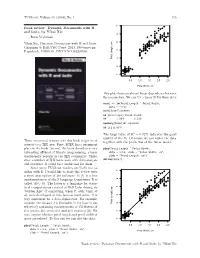
TUGboat, Volume 35 (2014), No. 1 115 ● ● ● Book review: Dynamic Documents with R ● ● ● and knitr, by Yihui Xie ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Boris Veytsman ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● Yihui Xie, Dynamic Documents with R and knitr. ● ● ● ● ● ● ● ● ● ● ● ● ● ● Chapman & Hall/CRC Press, 2013, 190+xxvi pp. ● ● ● ● ● ● ● ● ● US$ ISBN ● Paperback, 59.95. 978-1482203530. ● ● ● ● Petal Length, cm Petal ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● ● 1 2 3 4 5 6 7 0.5 1.0 1.5 2.0 2.5 Petal Width, cm This plot shows an almost linear dependence between the parameters. We can try a linear fit for these data: model <- lm(Petal.Length ˜ Petal.Width, data = iris) model$coefficients ## (Intercept) Petal.Width ## 1.084 2.230 summary(model)$r.squared ## [1] 0.9271 The large value of R2 = 0.9271 indicates the good quality of the fit. Of course we can replot the data There are several reasons why this book might be of together with the prediction of the linear model: interest to a TEX user. First, LATEX has a prominent place in the book. Second, the book describes a very plot(Petal.Length ˜ Petal.Width, interesting offshoot of literate programming, a topic data = iris, xlab = "Petal Width, cm", traditionally popular in the TEX community. Third, ylab = "Petal Length, cm") abline(model) since a number of TEX users work with data analysis and statistics, R could be a useful tool for them. Since some TUGboat readers are likely not fa- ● ● ● miliar with R, I would like to start this review with ● ● a short description of the software. R [1] is a free ● ● ● ● ● ● ● ● ● ● ● implementation of the S language (sometimes R is ● ● ● ● ● ● ● ● ● ● ● ● ● ● called GNU S). -

LYX and Knitr
knitr.1 LYX and knitr The knitr package allows one to embed R code within LYX and LATEX documents. When a document is compiled into a PDF, LYX/LATEX connects to R to run the R code and the code/output is automatically put into the PDF. In addition to this being a convenient way to use both LYX/LATEX with R, it also provides an important component to the reproducibility of research (RR). For example, one can include the code for a data analysis de- scribed in a paper. This ensures that there would be no “copying and pasting errors” and also provide readers of the paper an im- mediate way to reproduce the research. RR continues to become more important and fortunately more tools are being developed to make it possible. Below are some discussions on the topic: • AMSTAT News column on RR at http://magazine. amstat.org/blog/2011/01/01/scipolicyjan11. • CRAN task view for RR and R at http://cran.r-project. org/web/views/ReproducibleResearch.html. • Yihui Xie: Author of knitr – First and second editions of his Dynamic Documents with R and knitr book. Note that this book was typed in LYX. – Website for knitr at http://yihui.name/knitr The Sweave environment is another way to include R code inside of LYX/LATEX. This was developed prior to knitr, but it is more difficult to use. The purpose of this section is to examine the main compo- nents of knitr so that you will be able to complete the rest of the semester using LYX and knitr together for all assign- ments in our course! Also, a very important purpose is to give you the tools needed to complete all assignments in other R- based courses by using knitr and LYX together! The files used knitr.2 here are intro_example_cereal.lyx, intro_example_cereal.pdf, cereal.csv, ExternalCode.R, FirstBeamer-knitr.zip, JSM2015.zip, and RMarkdown.zip. -

Book of Abstracts
Book of Abstracts June 27, 2015 1 Conference Sponsors Diamond Sponsor Platinum Sponsors Gold Sponsors Silver Sponsors Open Analytics Bronze Sponsors Media Sponsors 2 Conference program Time Tuesday Wednesday Thursday Friday 08:00 Registration opens Registration opens Registration opens Registration opens 08:30 – 09:00 Opening session (by Rector peR! M. Johansen, Aalborg University) Aalborghallen 09:00 – 10:00 Romain François Di Cook Thomas Lumley Aalborghallen Aalborghallen Aalborghallen 10:00 – 10:30 Coffee break Coffee break Coffee break (15 min) ee break Sponsored by Quantide Sponsored by Alteryx ff Session 1 Session 4 10:30 – 12:00 Sponsor session (10:15) Kaleidoscope 1 Kaleidoscope 4 Aalborghallen Aalborghallen Aalborghallen incl. co Morning Tutorials DataRobot Ecology Medicine Gæstesalen Gæstesalen RStudio Teradata Networks Regression Musiksalen Musiksalen Revolution Analytics Reproducibility Commercial Offerings alteryx Det Lille Teater Det Lille Teater TIBCO H O Interfacing Interactive graphics 2 Radiosalen Radiosalen HP 12:00 – 13:00 Sandwiches Lunch (standing buffet) Lunch (standing buffet) Break: 12:00 – 12:30 Sponsored by Sponsored by TIBCO ff Revolution Analytics Ste en Lauritzen (12:30) Aalborghallen Session 2 Session 5 13:00 – 14:30 13:30: Closing remarks Kaleidoscope 2 Kaleidoscope 5 Aalborghallen Aalborghallen 13:45: Grab ’n go lunch 14:00: Conference ends Case study Teaching 1 Gæstesalen Gæstesalen Clustering Statistical Methodology 1 Musiksalen Musiksalen ee break Data Management Machine Learning 1 ff Det Lille Teater Det Lille -

The R Journal, June 2012
The Journal Volume 2/1, June 2010 A peer-reviewed, open-access publication of the R Foundation for Statistical Computing Contents Editorial..................................................3 Contributed Research Articles IsoGene: An R Package for Analyzing Dose-response Studies in Microarray Experiments..5 MCMC for Generalized Linear Mixed Models with glmmBUGS ................. 13 Mapping and Measuring Country Shapes............................... 18 tmvtnorm: A Package for the Truncated Multivariate Normal Distribution........... 25 neuralnet: Training of Neural Networks............................... 30 glmperm: A Permutation of Regressor Residuals Test for Inference in Generalized Linear Models.................................................. 39 Online Reproducible Research: An Application to Multivariate Analysis of Bacterial DNA Fingerprint Data............................................ 44 Two-sided Exact Tests and Matching Confidence Intervals for Discrete Data.......... 53 Book Reviews A Beginner’s Guide to R......................................... 59 News and Notes Conference Review: The 2nd Chinese R Conference........................ 60 Introducing NppToR: R Interaction for Notepad++......................... 62 Changes in R 2.10.1–2.11.1........................................ 64 Changes on CRAN............................................ 72 News from the Bioconductor Project.................................. 85 R Foundation News........................................... 86 2 The Journal is a peer-reviewed publication of -

Introduction to R: Part V Annex
Introduction to R: Part V Annex Alexandre Perera i Lluna1;2 1Centre de Recerca en Enginyeria Biomèdica (CREB) Departament d’Enginyeria de Sistemes, Automàtica i Informàtica Industrial (ESAII) Universitat Politècnica de Catalunya mailto:[email protected] 2Centro de Investigación Biomédica en Red en Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN) Jan 2011 / Introduction to R Universitat Rovira i Virgili Integration to other Languages Writing parallel code in R Bibliography ContentsI 1 Integration to other Languages R4Calc, OpenOffice LATEX R integration into Latex, Sweave 2 Writing parallel code in R Parallel code: when, how and why? R, Rmpi, Snow 3 Bibliography Alexandre Perera i Lluna; Introduction to R: Part V Integration to other Languages R4Calc, OpenOffice Writing parallel code in R LATEX Bibliography R integration into Latex, Sweave R and Calc R and Calc is an extension to Calc application, part of the OO.o suite (OpenOffice.org): Allows to see R objects from the OO.o menu R should be started with Rserve running: library(Rserve) Rserve() in OO.o, Use the menu bar R Add-on > Rdump() It will open a dialog, input rnorm(10) cor.test(c(1,2,3,4), c(1,2,2,3)) A new sheet will open with the result of the link to R http: //wiki.services.openoffice.org/wiki/R_and_Calc Alexandre Perera i Lluna; Introduction to R: Part V Integration to other Languages R4Calc, OpenOffice Writing parallel code in R LATEX Bibliography R integration into Latex, Sweave What is latex? LATEX (written as LaTeX in plain text, pronounced as /l\'atej/ ) is: Is a document markup language Is document preparation system for the TEX typesetting program Originally written in 1984 by Leslie Lamport at SRI International Only to the language in which documents are written, not to the text editor itself.