Developmental Genetics of Color Pattern Establishment in Cats ✉ Christopher B
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Savannah Cat’ ‘Savannah the Including Serval Hybrids Felis Catus (Domestic Cat), (Serval) and (Serval) Hybrids Of
Invasive animal risk assessment Biosecurity Queensland Agriculture Fisheries and Department of Serval hybrids Hybrids of Leptailurus serval (serval) and Felis catus (domestic cat), including the ‘savannah cat’ Anna Markula, Martin Hannan-Jones and Steve Csurhes First published 2009 Updated 2016 © State of Queensland, 2016. The Queensland Government supports and encourages the dissemination and exchange of its information. The copyright in this publication is licensed under a Creative Commons Attribution 3.0 Australia (CC BY) licence. You must keep intact the copyright notice and attribute the State of Queensland as the source of the publication. Note: Some content in this publication may have different licence terms as indicated. For more information on this licence visit http://creativecommons.org/licenses/ by/3.0/au/deed.en" http://creativecommons.org/licenses/by/3.0/au/deed.en Front cover: Close-up of a 4-month old F1 Savannah cat. Note the occelli on the back of the relaxed ears, and the tear-stain markings which run down the side of the nose. Photo: Jason Douglas. Image from Wikimedia Commons under a Public Domain Licence. Invasive animal risk assessment: Savannah cat Felis catus (hybrid of Leptailurus serval) 2 Contents Introduction 4 Identity of taxa under review 5 Identification of hybrids 8 Description 10 Biology 11 Life history 11 Savannah cat breed history 11 Behaviour 12 Diet 12 Predators and diseases 12 Legal status of serval hybrids including savannah cats (overseas) 13 Legal status of serval hybrids including savannah cats -

THE INTERNATIONAL CAT ASSOCIATION, INC. (Open Session
THE INTERNATIONAL CAT ASSOCIATION, INC. 2007 Annual Board Meeting August 29-31, 2007 Albuquerque, NM (Open Session) August 29, 2007, Wednesday, 8 AM ACTION TIME PAGE Welcome and Call to Order DeVilbiss Verbal 8-8:30AM - 1. Roll Call DeVilbiss Verbal - 2. President's Remarks DeVilbiss Verbal - (Executive Session) 8:30-11:30AM See Executive Agenda Lunch - 11:30AM-1PM (Open Session) Consent Agenda 1:00-1:15PM 1. Annual Awards Report EO Approve 5 2. License Fee Report EO Approve 7 3. Insurance Report EO Approve 9 4. Active Regional Clubs EO Approve Previously furnished - 5. Dom X Dom Litters Registered - None EO Approve - 6. Future Annuals, Semi-Annuals EO Approve 11 7. Minutes, Corrections/Additions EO Approve - 8. TICA TREND Editor Contract BOD Approve 12 9. Pet Pac Donation BOD Approve - 10. Reiss Design Assoc Agreement BOD Approve 15 Board Governance 1:15-2:00PM 1. Review Board Governance Policy Fisher Approve - a. Publish Minutes of the Meeting Fisher Add and Approve - 2. Review follow-up status report Fisher Approve 17 Fiduciary 2:00-3:00PM 1. 2007-2008 Budget Review EO Information 18 2. Overtime Report (End of Fiscal Year) EO Information 23 3. TDS Revenue FY 06-07 EO Information 24 4. Credit Card Info and new Addendum EO Approval 25 5. TICA TREND Final Report EO Information 32 Page 1 2007 Annual Meeting Agenda, Page 1 Discussions 3:00-5:00PM 1. Results-Russian Poll on Isolated Status Wood Discussion - 2. Results-American Bobtail Poll Crockett Discussion - 3. TICA FR CN club-NEW organization Christian Discussion - 4. -
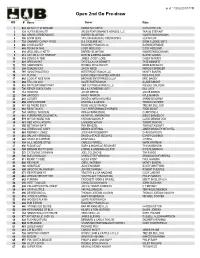
Open 2Nd Go Pre-Draw
as of: 11/23/2020 5:17 PM Open 2nd Go Pre-draw WO # Horse Owner Rider 1 543 METALLIC STARSHINE KENNETH FLIPPIN KEITH KITCHEN 2 135 AUTTA BE NAUTTI WEISS PERFORMANCE HORSES, LLC. TRAVIS STEWART 3 743 SENSEUVEBEENGONE DARREN BLANTON KAMERON BUCHANAN 4 186 BOON JEAN TAYLOR RANCHING OPERATIONS HEATH COX 5 732 SANNREY CATNIP SPICE D & S EQUINE INC DENNIS (ZEKE) ENTZ 6 496 LOUELLA REY ROCKING P RANCH LLC GLEN BEVERIDGE 7 889 TEXAS IN 1880 CODY HEDLUND CODY HEDLUND 8 934 WELL HELLO KITTY DARREN BLANTON KAMERON BUCHANAN 9 219 CAT ON A HOT TINROOF DUSTIN & DEENA ADAMS DUSTIN ADAMS 10 315 ECHOS IN TIME JAMES (CODY) LORE CASEY MORRIS 11 821 SPECIALKAT TATE & LAURA BENNETT TATE BENNETT 12 701 SABIDOREYA DOUBLE DOVE RANCH ADAN BANUELOS 13 309 DUALINS CAT JASON REED MARCUS SPENCER 15 797 SMOOTH MATRIXX BITTERROOT RANCH LLC MONTY BUNTIN 16 337 FLYISH BUCK CREEK QUARTER HORSES WES ASHLOCK 17 489 LOOK AT MEE NOW BROWNE ENTERPRISES LLLP ERIC MAGBY 18 888 TELL ON LOU SLATE RIVER RANCH GLADE KNIGHT 19 653 RATTLERS BABY PHAT SMF CUTTING HORSES LLC WESLEY GALYEAN 20 700 REYZN QUICK CASH BILL & CATHERINE LACY BILL LACY 21 712 ROCKITS JACOB MEEKS JACOB MEEKS 22 784 SMOOCCI HAVEY MANION KYLE MANION 23 248 CLOSER DAVID & MARLA HUGHES SHANE BINGHAM 24 459 LEWS CHROME LEW HALL & ASSOC THOMAS SCHEER 25 147 BE THERE SUEN ROSE VALLEY RANCH TREVER BULLOCK 26 682 REYS TALKIN T & K PERFORMANCE HORSES TODD ADOLF 27 724 SADDLE THIS DEAL ORA & FRANK DIEHL CURRY DIEHL 28 643 PURRN PRECIOUS METAL KATHRYN LYNN BROWN DIEGO BANUELOS 29 579 MY WAYWARD SON CROWN RANCH LP LLOYD -

Guidance for USDA Accredited Veterinarians Preparing Cats for Export from the United States (Except Hawaii and Guam) to Australia
Guidance for USDA accredited veterinarians preparing cats for export from the United States (except Hawaii and Guam) to Australia • Cats traveling from the United States to Australia must be accompanied by a valid import permit, which provides the conditions for importing the cat. • Failure to comply with Australia’s import conditions, exactly as specified, may result in the cat being (at the owner’s cost): held longer in post entry quarantine, subject to additional testing, re- exported, or euthanized. • Read the cat’s import permit. • Follow the step-by-step guide. • Scan and verify the animal’s microchip every time it is tested, treated or examined prior to export. • Accurately record the animal’s microchip number on every document associated with the animal including any: o treatment, vaccination, and examination certificates o laboratory submission forms which accompany blood samples for testing • Only administer compliant external parasite treatments. Record the product name and active ingredient(s) on the health certificate. o A list (not exhaustive) of compliant external parasite treatments can be found here. o External parasite treatments must be topical (not oral) and kill ticks and fleas on contact. o Bravecto® (fluralaner), NexGard® (afoxolaner) and Revolution® (selamectin) are not compliant external parasite treatments. • Re-administer external parasite treatments in accordance with the manufacturer’s requirements so that coverage is continuous until export. If the product needs to be applied monthly, it must be applied no later than the 31st day from the last application. • Administer internal parasite treatments that are effective against both nematodes (roundworms) and cestodes (tapeworms). The animal must be treated twice before export. -
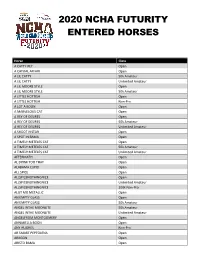
2020 Ncha Futurity Entered Horses
2020 NCHA FUTURITY ENTERED HORSES Horse Class A CATTY REY Open A CATUAL AFFAIR Open A LIL CATTY 50k Amateur A LIL CATTY Unlimited Amateur A LIL MOORE STYLE Open A LIL MOORE STYLE 50k Amateur A LITTLE HOTTISH Open A LITTLE HOTTISH Non-Pro A LOT A BOON Open A MARVELOUS CAT Open A REY OF DESIRES Open A REY OF DESIRES 50k Amateur A REY OF DESIRES Unlimited Amateur A SHOOT N STAR Open A SPOT IN BAMA Open A TIMELY METELES CAT Open A TIMELY METELES CAT 50k Amateur A TIMELY METELES CAT Unlimited Amateur AFTERMATH Open AL DRINK TOO THAT Open ALABAMA CUPID Open ALL SPICE Open ALLSPICENOTHINGNICE Open ALLSPICENOTHINGNICE Unlimited Amateur ALLSPICENOTHINGNICE 200k Non-Pro ALOT MO METALLIC Open AN EMPTY GLASS Open AN EMPTY GLASS 50k Amateur ANGEL INTHE MOONLITE 50k Amateur ANGEL INTHE MOONLITE Unlimited Amateur ANGELFROM MONTGOMERY Open ANNABELLA BOON Open ANY HUGHES Non-Pro AR SMART PEPTOLENA Open ARAGON Open ARISTO BAMA Open ARISTO BAMA Non-Pro ARISTO SALLY Open ASPENGLOW Open ASTURIA Unlimited Amateur ASTURIA 200k Non-Pro ATHENA BOON Open ATHENA CAT A REY Open AUGUSTUS MCCRAY MC Open AUGUSTUS METALLICA Open AUTTA BE NAUTTI Open AUTUM LILLY Open AUTUMN BE HIGH CLASS 200k Non-Pro AWESOME AUTUMNS CAT Open AWESOME N BLACK 50k Amateur AWESOME N BLACK Unlimited Amateur BABY BOONERS Open BABY TALK Open BACARDEE Open BACARDEE Non-Pro BACK ATCHA BABY Open BAD AND BOUJEE 50k Amateur BAD AND BOUJEE Unlimited Amateur BADABOOMSMALL Open BAMA MARJORETTE Open BAMA STATE OF MIND Open BAMBI KITTY Open BARR DOWN Non-Pro BARR DOWN Unlimited Amateur BE THERE SUEN -

Cat Breeder Directory CAT FANCY, P.O
T CAT FANCY FANCY CAT EXPLAINED BEHAVIOR • STRANGE & FUN HOLIDAY OF 2012 • SAFE NEW PRODUCTS • SINGAPURA BEST SNOWSHOE DECEMBER 2012 H A DECEMBER 2012 FREE POSTER! R E ® U S W P A E G E N T, SI * PETITE CAT FANCY CAT THE AUTHORITY ON ALL THINGS CAT ® EDITORS’ CHOICE: BEST New 23Products OF 2012 ➻ Strange Behavior EXPLAINED HOW MANY Words CAN YOUR CAT Learn? Make this Holiday Safe&Fun! Happy Kitty Holiday + CONTEST WINNERS Snowshoe The Ultimate People Cat CCFcover1212.inddFcover1212.indd 1 99/24/12/24/12 88:33:09:33:09 AAMM C2_C3_C4_CF1212 9/21/12 9:40 AM Page Cov2 Born to climb Born to stalk Born to pounce Did you know the African Wildcat is an original ancestor of your domestic cat? At Purina ONE, we believe that by better understanding the behavior and needs of the African Wildcat, we can understand who our cats were born to be and deepen our relationship with them. Why do cats jump? Pounce? Stalk? Join our journey online to learn more. > www.purinaone.com/borntobe Discover your cat’s true nature. ® ® All trademarks are owned by Société des Produits Nestlé S.A. or used with permission. Printed in U.S.A. 1EdNote1212 9/21/12 9:55 AM Page 1 FROM THE EDITOR VOLUME 55 • NUMBER 12 DECEMBER 2012 Holiday Miracles Editor Susan Logan Managing Editor Annie B. Shirreffs Managing Web Editor Anastasia Thrift HOW DOES A DAYS-OLD KITTEN SURVIVE GETTING SWEPT Art Director Jerome Callens up by a tornado and flung onto the top of a tree? Pre-weaned kittens Group Editor Ernie Slone Web Editorial Director require so much around-the-clock care that it doesn’t seem possible Melissa Kauffman that one, aptly named Toto, could survive that. -

Savannahs Do Make Great Pets, Don't Believe BCR's Hype
Savannahs Do Make Great Pets, Don’t believe BCR’s Hype. By Brigitte Cowell, Ph.D., Kirembo Savannahs, www.kirembosavannahs.com Rescue Coordinator for Savannah Rescue. I’ve been a cat breeder for a few years now. I breed Savannah cats, an exciting new domestic breed of cat accepted by the International Cat Association (TICA) and one of their fastest growing new breeds. Not only do I spend my days promoting this highly exciting new cat breed, but I also head up the breed Rescue for Savannah cats along with my colleague Kristine Alessio. Big Cat Rescue is responsible for a great deal of misinformation about domestic hybrid cat breeds such as Bengals, Chausies and Savannahs. As I deal mainly with Savannahs, although I have fostered Chausies and Bengals in the past. Most of my comments below are taken from my experience with that particular domestic cat breed. Big Cat Rescue, BCR, is wrong about many things. The error they are making that I currently object to is that they are labeling the Savannah as an “exotic”; so right in the same grouping as lions and tigers is a domestic cat breed recognized by the largest registry of DOMESTIC cats, (TICA, the International Cat Association). According to BCR, you should put a domestic Savannah cat in a cage instead of your home. USDA defines the Savannah as domestic* because it always has a domestic parent even at the first generation. Savannahs are a domestic pet: they eat domestic cat food, they use litter boxes, play with cat toys and sleep on your bed. -

THE INTERNATIONAL CAT ASSOCIATION, INC. 2008 Annual Board Meeting August 27-29, 2008 Arlington, TX
THE INTERNATIONAL CAT ASSOCIATION, INC. 2008 Annual Board Meeting August 27-29, 2008 Arlington, TX The meeting was called to order at 8:45AM, by the President, Kay DeVillbiss. The following Board members were present: Kay DeVilbiss, President Ellen Crockett, Northwest Motoko Oizumi, Asia Carlos Arrieta, South America Jamie Christian, Great Lakes Cheryl Hogan, South Central Linda Kay Ashley, Mid Paciific Jo Parris, Southeast Lisa Dickie, Mid Atlantic Vickie Fisher, Southwest Martin Wood, Northern Europe Genevieve Basquine, Southern Europe Donna Madison, Northeast The Vice President, Nancy Parkinson was unable to attend. The following members were also in attendance: Dewane Barnes, Rules Chair/Parliamentarian Laurie Schiff, Legal Advisor Bobbie Tullo, Judging Administrator Micki Takei Landa, Interpretor for Asia Frances Young, Legal Advisor The President, when presiding over the meeting, refrains from voting except when the vote is by ballot, or whenever her vote will affect the result. 1. Unanimous consent to go into Executive Session. Without objection. 2. Unanimous consent to allow the Interpreter, Judging Administrator , Rules Chair and the attorneys to remain in Executive Session. Without objection. 3. Motion was made by Parris and seconded by Ashley to contact Dave Clark’s attorney to propose a settlement which would prohibit Clark from verbally communicating with the Executive Office for 5 years. Motion carried unanimously. 4. Motion was made by Parris and seconded by Fisher to ask the Judging Administrator to contact Barrett. Motion carried with Madison opposed and Basquine and Oizumi abstaining. 5. Motion was made by Christian and seconded by Hogan to set a hearing on the Dany v Verbeeren matter. -

Photo Gallery of Wild and Exotic Cats and Their Hybrids
REXANO, 205 N. Stephanie Street, Suite D # 131, Henderson, Nevada 89074 Photo Gallery of Wild and Exotic Cats and their Hybrids The last few years has brought an increase in bans on the private ownership of wild and exotic cats in the USA. What most legislators or even the general public doesn’t realize is that the majority of wildcats and their hybrids are small and don’t pose any more threat or need for recapture equipment than the feral cats. To the average person or most animal control officers, many are undistinguishable from an alley cat. The current trend is “fear of the unknown”; if it sounds exotic, then it needs to be banned. We are hoping to reverse this trend with this educational flyer on wild and exotic cats. SMALL WILD AND EXOTIC CATS (Partial list) Far left: Black- Footed Cat (Felis nigripes) Weight Females 3 pounds Males 4 pounds Left: Kodkod, Guiña (Leopardus guigna) Weight 4-5.5 pounds Sand Cat (Felis margarita) Male weight:- 5-7 pounds Female weight: 3- 5 pounds Geoffroy's Cat (Leopardus geoffroyi) Weight: 4-12 pounds Melanistic Geoffroy's Cat (Leopardus geoffroyi) Weight: 4-12 pounds Jaguarundi (Puma yagouaroundi) Weight up to 14 pounds Asian Leopard Cat, ALC (Prionailurus bengalensis) Weight: 10-15 pounds Margay (Leopardus wiedii) Weight: 6-20 pounds Far left:Wild cat (Felis silvestris), Pictured: Gordon's Cat (Felis silvestris gordoni) Up to 18 pounds Left: Fishing Cat (Prionailurus viverrinus) Up to 26 pounds Jungle cat (Felis chaus) Weight 9-35 pounds Bobcat (Lynx rufus) Weight: males 16 to 35 pounds, females -

Guide to Importing Dogs and Cats Into Japan from Non-Designated Regions (Final Revision: Mar., 2021)
Guide to importing dogs and cats (from Non-designated regions) Appended Form 5-2 Guide to importing dogs and cats into Japan from Non-designated regions (Final revision: Mar., 2021) Animal Quarantine Service Ministry of Agriculture, Forestry and Fisheries What are the “Non-designated regions”? All countries or regions other than Iceland, Australia, New Zealand, Fiji Islands, Hawaii and Guam. *The designation is subject to change due to the prevalence of rabies or the revision of quarantine regulations. This guide describes the requirements and necessary procedures for importing dogs and cats into Japan from Non-designated regions. We strongly recommend reading this guide if you are planning to bring dogs or cats into Japan. For further information, feel free to contact Animal Quarantine Service. 1 Guide to importing dogs and cats (from Non-designated regions) Contents A. Introduction ........................................................................................................................ 3 Importer’s responsibility................................................................................................... 3 Animals covered by this guide ......................................................................................... 3 B. Import quarantine procedures ............................................................................................ 4 Flow chart .......................................................................................................................... 4 STEP 1: Microchip ............................................................................................................ -

Vaccines for Savannah Cats
Vaccines for Savannah Cats Before we go any further, Savannah Cats do not need ‘special’ vaccines. There is an old-wives- tale in the Savannah cat community that Savannah cats should never have Modified Live Virus (MLV) vaccines. This is simply not true. Many of us even give MLV vaccines to our serval cats. That being said, let’s discuss what a vaccination is and why a responsible breeder pays so much attention to vaccinating. Laboratories that make Vaccines Various laboratories prepare vaccines. The labs take very small amounts of the virus or bacteria it will protect against and modify it to form vaccines. They then distribute it in small, sterile bottles. These are for sale to vets and breeders to administer to cats and kittens. Thie vaccines increase immunity in our cats, from a specific disease or virus. The two types of vaccines, killed and MLV, work in different ways. Rabies There are no general laws, except for rabies, that says a cat/kitten must have a periodic vaccine. Even now some vaccine manufacturers are making a three-year vaccine. More people are beginning to realize that following the practice of a yearly vaccine that our veterinarians recommend is over-vaccinating and we must weigh the benefits and risks. There are studies now where older cats 14+ years have had titer* tests run and have antibodies from their original kitten vaccines. Adjuvants are the Main Cause of Injection Site Sarcomas Many of the killed vaccines contain adjuvants. Adjuvants, defined by Wikipedia is an immunological agent that modifies the effect of other agents. -

Savannah Cat (Leptailurus Serval X Felis Catus)
Indicative 10 Project National Resource Material Savannah Cat (Leptailurus serval x Felis catus) Win Kirkpatrick and Michelle T. Christy 2017 Department of Primary Industries and Regional Development 3 Baron-Hay Court, South Perth, WA 6151 An Invasive Animals CRC Project Contents Summary ...................................................................................... 1 Key Messages ............................................................................ 1 Classification ............................................................................ 1 Common names ......................................................................... 1 Biology and Ecology ......................................................................... 2 Identification ............................................................................ 2 Behaviours and Traits.................................................................. 3 Food and Foraging ...................................................................... 4 Reproduction and Lifecycle .......................................................... 4 Global Range ................................................................................. 4 Potential for Introduction ................................................................. 4 Potential for Eradication .................................................................. 5 Impacts ........................................................................................ 5 Economic ................................................................................