Overview of the Most Prevalent Hypothalamus-Specific Mrnas, As Identified by Directional Tag PCR Subtraction KAARE M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Role of the Hypothalamic Dorsomedial Nucleus in the Central Regulation of Food Intake
The role of the hypothalamic dorsomedial nucleus in the central regulation of food intake Ph.D. thesis Éva Dobolyiné Renner Semmelweis University Szentágothai János Neuroscience Doctoral School Ph.D. Supervisor: Prof. Miklós Palkovits, member of the HAS Opponents: Prof. Dr. Halasy Katalin, Ph.D., D.Sci. Dr. Oláh Márk, M.D., Ph.D. Examination board: Prof. Dr. Vígh Béla, M.D., Ph.D., D.Sci Dr. Kovács Krisztina Judit, Ph.D., D.Sci. Dr. Lovas Gábor, M.D., Ph.D. Budapest 2013 1. Introduction The central role of the hypothalamus in the regulation of food intake and energy expenditure has long been established. The hypothalamus receives hormonal input such as insulin, leptin, and ghrelin from the periphery. The gate for the most important adiposity signals is the arcuate nucleus, which contains neurons expressing orexigenic and anorexigenic peptides, respectively. These neurons convey peripheral input to the paraventricular and ventromedial nuclei, and the lateral hypothalamic area, which all play critical roles in body weight regulations. The hypothalamic dorsomedial nucleus (DMH) has also been implicated in the regulation of body weight homeostasis along with other hypothalamic nuclei including the arcuate, ventromedial, and paraventricular nuclei as well as the lateral hypothalamus. Lesions of the DMH affected ingestive behavior. Electrophysiological data suggested that neurons in this nucleus integrate hormonal input and ascending brainstem information and, in turn, modulate food intake and energy balance. In response to refeeding of fasted rats, Fos-activated neurons were reported in the DMH. Major projections relay vagus-mediated signals from the gastrointestinal tract, and humoral signals to the hypothalamus from the nucleus of the solitary tract (NTS), a viscerosensory cell group in the dorsomedial medulla. -

Cortex and Thalamus Lecture.Pptx
Cerebral Cortex and Thalamus Hyperbrain Ch 2 Monica Vetter, PhD January 24, 2013 Learning Objectives: • Anatomy of the lobes of the cortex • Relationship of thalamus to cortex • Layers and connectivity of the cortex • Vascular supply to cortex • Understand the location and function of hypothalamus and pituitary • Anatomy of the basal ganglia • Primary functions of the different lobes/ cortical regions – neurological findings 1 Types of Cortex • Sensory (Primary) • Motor (Primary) • Unimodal association • Multimodal association - necessary for language, reason, plan, imagine, create Note: • Gyri • Sulci • Fissures • Lobes 2 The Thalamus is highly interconnected with the cerebral cortex, and handles most information traveling to or from the cortex. “Specific thalamic Ignore nuclei” – have well- names of defined sensory or thalamic nuclei for motor functions now - A few Other nuclei have will more distributed reappear later function 3 Thalamus Midbrain Pons Limbic lobe = cingulate gyrus Structure of Neocortex (6 layers) white matter gray matter Pyramidal cells 4 Connectivity of neurons in different cortical layers Afferents = inputs Efferents = outputs (reciprocal) brainstem etc Eg. Motor – Eg. Sensory – more efferent more afferent output input Cortico- cortical From Thalamus To spinal cord, brainstem etc. To Thalamus Afferent and efferent connections to different ….Depending on whether they have more layers of cortex afferent or efferent connections 5 Different areas of cortex were defined by differences in layer thickness, and size and -

Critical Role of Dorsomedial Hypothalamic Nucleus in a Wide Range of Behavioral Circadian Rhythms
The Journal of Neuroscience, November 19, 2003 • 23(33):10691–10702 • 10691 Behavioral/Systems/Cognitive Critical Role of Dorsomedial Hypothalamic Nucleus in a Wide Range of Behavioral Circadian Rhythms Thomas C. Chou,1,2 Thomas E. Scammell,2 Joshua J. Gooley,1,2 Stephanie E. Gaus,1,2 Clifford B. Saper,2 and Jun Lu2 1Department of Neurobiology and Program in Neuroscience, Harvard Medical School, Boston, Massachusetts 02115, and 2Department of Neurology, Harvard Medical School and Beth Israel Deaconess Medical Center, Boston, Massachusetts 02215 The suprachiasmatic nucleus (SCN) contains the brain’s circadian pacemaker, but mechanisms by which it controls circadian rhythms of sleep and related behaviors are poorly understood. Previous anatomic evidence has implicated the dorsomedial hypothalamic nucleus (DMH) in circadian control of sleep, but this hypothesis remains untested. We now show that excitotoxic lesions of the DMH reduce circadian rhythms of wakefulness, feeding, locomotor activity, and serum corticosteroid levels by 78–89% while also reducing their overall daily levels. We also show that the DMH receives both direct and indirect SCN inputs and sends a mainly GABAergic projection to the sleep-promoting ventrolateral preoptic nucleus, and a mainly glutamate–thyrotropin-releasing hormone projection to the wake- promoting lateral hypothalamic area, including orexin (hypocretin) neurons. Through these pathways, the DMH may influence a wide range of behavioral circadian rhythms. Key words: suprachiasmatic nucleus; sleep; feeding; corticosteroid; locomotor activity; melatonin Introduction sponses, in feeding, and in the circadian release of corticosteroids In many animals, the suprachiasmatic nucleus (SCN) strongly (for review, see Bellinger et al., 1976; Kalsbeek et al., 1996; Ber- influences circadian rhythms of sleep–wake behaviors, but the nardis and Bellinger, 1998; DiMicco et al., 2002), although many pathways mediating these influences are poorly understood. -

The Human Thalamus Is an Integrative Hub for Functional Brain Networks
5594 • The Journal of Neuroscience, June 7, 2017 • 37(23):5594–5607 Behavioral/Cognitive The Human Thalamus Is an Integrative Hub for Functional Brain Networks X Kai Hwang, Maxwell A. Bertolero, XWilliam B. Liu, and XMark D’Esposito Helen Wills Neuroscience Institute and Department of Psychology, University of California, Berkeley, Berkeley, California 94720 The thalamus is globally connected with distributed cortical regions, yet the functional significance of this extensive thalamocortical connectivityremainslargelyunknown.Byperforminggraph-theoreticanalysesonthalamocorticalfunctionalconnectivitydatacollected from human participants, we found that most thalamic subdivisions display network properties that are capable of integrating multi- modal information across diverse cortical functional networks. From a meta-analysis of a large dataset of functional brain-imaging experiments, we further found that the thalamus is involved in multiple cognitive functions. Finally, we found that focal thalamic lesions in humans have widespread distal effects, disrupting the modular organization of cortical functional networks. This converging evidence suggests that the human thalamus is a critical hub region that could integrate diverse information being processed throughout the cerebral cortex as well as maintain the modular structure of cortical functional networks. Key words: brain networks; diaschisis; functional connectivity; graph theory; thalamus Significance Statement The thalamus is traditionally viewed as a passive relay station of information from sensory organs or subcortical structures to the cortex. However, the thalamus has extensive connections with the entire cerebral cortex, which can also serve to integrate infor- mation processing between cortical regions. In this study, we demonstrate that multiple thalamic subdivisions display network properties that are capable of integrating information across multiple functional brain networks. Moreover, the thalamus is engaged by tasks requiring multiple cognitive functions. -

Basal Ganglia Anatomy, Physiology, and Function Ns201c
Basal Ganglia Anatomy, Physiology, and Function NS201c Human Basal Ganglia Anatomy Basal Ganglia Circuits: The ‘Classical’ Model of Direct and Indirect Pathway Function Motor Cortex Premotor Cortex + Glutamate Striatum GPe GPi/SNr Dopamine + - GABA - Motor Thalamus SNc STN Analagous rodent basal ganglia nuclei Gross anatomy of the striatum: gateway to the basal ganglia rodent Dorsomedial striatum: -Inputs predominantly from mPFC, thalamus, VTA Dorsolateral striatum: -Inputs from sensorimotor cortex, thalamus, SNc Ventral striatum: Striatal subregions: Dorsomedial (caudate) -Inputs from vPFC, hippocampus, amygdala, Dorsolateral (putamen) thalamus, VTA Ventral (nucleus accumbens) Gross anatomy of the striatum: patch and matrix compartments Patch/Striosome: -substance P -mu-opioid receptor Matrix: -ChAT and AChE -somatostatin Microanatomy of the striatum: cell types Projection neurons: MSN: medium spiny neuron (GABA) •striatonigral projecting – ‘direct pathway’ •striatopallidal projecting – ‘indirect pathway’ Interneurons: FS: fast-spiking interneuron (GABA) LTS: low-threshold spiking interneuron (GABA) LA: large aspiny neuron (ACh) 30 um Cellular properties of striatal neurons Microanatomy of the striatum: striatal microcircuits • Feedforward inhibition (mediated by fast-spiking interneurons) • Lateral feedback inhibition (mediated by MSN collaterals) Basal Ganglia Circuits: The ‘Classical’ Model of Direct and Indirect Pathway Function Motor Cortex Premotor Cortex + Glutamate Striatum GPe GPi/SNr Dopamine + - GABA - Motor Thalamus SNc STN The simplified ‘classical’ model of basal ganglia circuit function • Information encoded as firing rate • Basal ganglia circuit is linear and unidirectional • Dopamine exerts opposing effects on direct and indirect pathway MSNs Basal ganglia motor circuit: direct pathway Motor Cortex Premotor Cortex Glutamate Striatum GPe GPi/SNr Dopamine + GABA Motor Thalamus SNc STN Direct pathway MSNs express: D1, M4 receptors, Sub. -

Aversive Stimuli Drive Hypothalamus-To-Habenula
Aversive stimuli drive hypothalamus-to-habenula excitation to promote escape behavior Salvatore Lecca, Frank Meye, Massimo Trusel, Anna Tchenio, Julia Harris, Martin Karl Schwarz, Denis Burdakov, F Georges, Manuel Mameli To cite this version: Salvatore Lecca, Frank Meye, Massimo Trusel, Anna Tchenio, Julia Harris, et al.. Aversive stimuli drive hypothalamus-to-habenula excitation to promote escape behavior. eLife, eLife Sciences Publication, 2017, 6, pp.e30697. 10.7554/eLife.30697. hal-03129952 HAL Id: hal-03129952 https://hal.archives-ouvertes.fr/hal-03129952 Submitted on 3 Feb 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. SHORT REPORT Aversive stimuli drive hypothalamus-to- habenula excitation to promote escape behavior Salvatore Lecca1,2, Frank Julius Meye1,3, Massimo Trusel1,2, Anna Tchenio1,2, Julia Harris4, Martin Karl Schwarz5, Denis Burdakov4, Francois Georges6,7, Manuel Mameli1,2* 1Institut du Fer a` Moulin, Inserm UMR-S 839, Paris, France; 2Department of Fundamental Neuroscience, The University of Lausanne, Lausanne, Switzerland; -

The Thalamus: Gateway to the Mind Lawrence M
Overview The thalamus: gateway to the mind Lawrence M. Ward∗ The thalamus of the brain is far more than the simple sensory relay it was long thought to be. From its location at the top of the brain stem it interacts directly with nearly every part of the brain. Its dense loops into and out of cortex render it functionally a seventh cortical layer. Moreover, it receives and sends connections to most subcortical areas as well. Of course it does function as a very sophisticated sensory relay and thus is of vital importance to perception. But also it functions critically in all mental operations, including attention, memory, and consciousness, likely in different ways for different processes, as indicated by the consequences of damage to its various nuclei as well as by invasive studies in nonhuman animals. It plays a critical role also in the arousal system of the brain, in emotion, in movement, and in coordinating cortical computations. Given these important functional roles, and the dearth of knowledge about the details of its nonsensory nuclei, it is an attractive target for intensive study in the future, particularly in regard to its role in healthy and impaired cognitive functioning. © 2013 John Wiley & Sons, Ltd. How to cite this article: WIREs Cogn Sci 2013. doi: 10.1002/wcs.1256 INTRODUCTION and other animals can do quite well without major chunks of cortex. Indeed, decorticate rats behave very pen nearly any textbook of neuroscience or similarly to normal rats in many ways,2 whereas sensation and perception and you will find the O de-thalamate rats die. -

Tonic Activity in Lateral Habenula Neurons Promotes Disengagement from Reward-Seeking Behavior
bioRxiv preprint doi: https://doi.org/10.1101/2021.01.15.426914; this version posted January 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Tonic activity in lateral habenula neurons promotes disengagement from reward-seeking behavior 5 Brianna J. Sleezer1,3, Ryan J. Post1,2,3, David A. Bulkin1,2,3, R. Becket Ebitz4, Vladlena Lee1, Kasey Han1, Melissa R. Warden1,2,5,* 1Department of Neurobiology and Behavior, Cornell University, Ithaca, NY 14853, USA 2Cornell Neurotech, Cornell University, Ithaca, NY 14853 USA 10 3These authors contributed equally 4Department oF Neuroscience, Université de Montréal, Montréal, QC H3T 1J4, Canada 5Lead Contact *Correspondence: [email protected] 15 Sleezer*, Post*, Bulkin* et al. 1 of 38 bioRxiv preprint doi: https://doi.org/10.1101/2021.01.15.426914; this version posted January 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. SUMMARY Survival requires both the ability to persistently pursue goals and the ability to determine when it is time to stop, an adaptive balance of perseverance and disengagement. Neural activity in the 5 lateral habenula (LHb) has been linked to aversion and negative valence, but its role in regulating the balance between reward-seeking and disengaged behavioral states remains unclear. -

Hypothalamus and Pituitary Gland Development in the Common Snapping Turtle, Chelydra Serpentina, and Disruption with Atrazine Exposure
University of North Dakota UND Scholarly Commons Theses and Dissertations Theses, Dissertations, and Senior Projects January 2016 Hypothalamus And Pituitary Gland Development In The ommonC Snapping Turtle, Chelydra Serpentina, And Disruption With Atrazine Exposure Kathryn Lee Gruchalla Russart Follow this and additional works at: https://commons.und.edu/theses Recommended Citation Russart, Kathryn Lee Gruchalla, "Hypothalamus And Pituitary Gland Development In The ommonC Snapping Turtle, Chelydra Serpentina, And Disruption With Atrazine Exposure" (2016). Theses and Dissertations. 2069. https://commons.und.edu/theses/2069 This Dissertation is brought to you for free and open access by the Theses, Dissertations, and Senior Projects at UND Scholarly Commons. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of UND Scholarly Commons. For more information, please contact [email protected]. HYPOTHALAMUS AND PITUITARY GLAND DEVELOPMENT IN THE COMMON SNAPPING TURTLE, CHELYDRA SERPENTINA, AND DISRUPTION WITH ATRAZINE EXPOSURE by Kathryn Lee Gruchalla Russart Bachelor of Science, Minnesota State University, Mankato, 2006 A Dissertation Submitted to the Graduate Faculty of the University of North Dakota in partial fulfillment of the requirements for the degree of Doctor of Philosophy Grand Forks, North Dakota August 2016 Copyright 2016 Kathryn Russart ii iii PERMISSION Title Hypothalamus and Pituitary Gland Development in the Common Snapping Turtle, Chelydra serpentina, and Disruption with Atrazine Exposure Department Biology Degree Doctor of Philosophy In presenting this dissertation in partial fulfillment of the requirements for a graduate degree from the University of North Dakota, I agree that the library of this University shall make it freely available for inspection. -
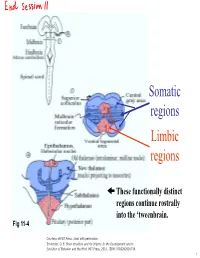
Lecture 12 Notes
Somatic regions Limbic regions These functionally distinct regions continue rostrally into the ‘tweenbrain. Fig 11-4 Courtesy of MIT Press. Used with permission. Schneider, G. E. Brain structure and its Origins: In the Development and in Evolution of Behavior and the Mind. MIT Press, 2014. ISBN: 9780262026734. 1 Chapter 11, questions about the somatic regions: 4) There are motor neurons located in the midbrain. What movements do those motor neurons control? (These direct outputs of the midbrain are not a subject of much discussion in the chapter.) 5) At the base of the midbrain (ventral side) one finds a fiber bundle that shows great differences in relative size in different species. Give examples. What are the fibers called and where do they originate? 8) A decussating group of axons called the brachium conjunctivum also varies greatly in size in different species. It is largest in species with the largest neocortex but does not come from the neocortex. From which structure does it come? Where does it terminate? (Try to guess before you look it up.) 2 Motor neurons of the midbrain that control somatic muscles: the oculomotor nuclei of cranial nerves III and IV. At this level, the oculomotor nucleus of nerve III is present. Fibers from retina to Superior Colliculus Brachium of Inferior Colliculus (auditory pathway to thalamus, also to SC) Oculomotor nucleus Spinothalamic tract (somatosensory; some fibers terminate in SC) Medial lemniscus Cerebral peduncle: contains Red corticospinal + corticopontine fibers, + cortex to hindbrain fibers nucleus (n. ruber) Tectospinal tract Rubrospinal tract Courtesy of MIT Press. Used with permission. Schneider, G. -

Neuromodulation in Primary Headaches
Neuromodulation in Primary Headaches Definition Neuromodulatory approaches can be divided into invasive procedures (peripheral nerve stimulation, vagal nerve stimulation, cervical spinal cord stimulation, and hypothalamic deep brain stimulation) and noninvasive procedures (transcutaneous electrical nerve stimulation [TENS] and transcranial magnetic and direct current stimulation). The underlying principle is a modulation of neuronal structures that are directly or indirectly involved in detection or transmission of painful stimuli or in the processing of this information in the brain. This approach comprises a direct modulation of brain structures involved in the generation of attacks (deep brain stimulation of the hypothalamus in cluster headache), modulation of inhibitory antinociceptive pathways (occipital nerve stimulation), modulation of cortical excitability (transcranial magnetic and direct current stimulation), and direct inhibitory effects at the level of the peripheral neuron or the spinal cord (TENS). Patient Selection While noninvasive techniques can be used widely, patients scheduled for invasive approaches should be selected carefully, since these techniques are still experimental and harbor potential hazards. Based on previously published consensus criteria for the definition of refractory chronic cluster headache (CCH) and chronic migraine, patients undergoing implantation of an invasive neuromodulatory approach should satisfy the following criteria: • The headache should be chronic and should have lasted for 2 years. • Established prophylactic drugs should have been tried without success (or were either not tolerated or contraindicated) in a sufficient dose for a sufficiently long period as monotherapy or combination therapy. In CCH, at least verapamil, topiramate, and lithium and in chronic migraine at least beta blockers, calcium antagonists, and anticonvulsants should have been tried. • Medication overuse should have been ruled out. -

Hypothalamushypothalamus -- Pituitarypituitary -- Adrenaladrenal Glandsglands
HypothalamusHypothalamus -- pituitarypituitary -- adrenaladrenal glandsglands Magdalena Gibas-Dorna MD, PhD Dept. of Physiology University of Medical Sciences Poznań, Poland Hypothalamus - general director of the hormone system. At every moment, the hypothalamus analyses messages coming from: the brain and different regions of the body. Homeostatic functions of hypothalamus include maintaining a stable body temperature, controlling food intake, controlling blood pressure, ensuring a fluid balance, and even proper sleep patterns. Cell bodies of neurons that produce releasing/inhibiting hormones Hypothalamus HypothalamusHypothalamus releases Arterial flow Primary capillaries in median eminence hormones at Long Releasing Portal hormones Anterior veins median eminence pituitary hormone Releasing/ inhibiting hormones and sends to anterior pituitary ANTERIOR PITUITARY via portalportal veinvein. Secretory cells that produce anterior pituitary hormones Anterior pituitary hormones Venous outflow Gonadotropic Thyroid- Proactin hormones stimulating ACTH Growth (FSH and LH) hormone hormone ControlControl ofof pituitarypituitary hormonehormone secretionsecretion byby hypothalamushypothalamus • Secretion by the anterioranterior pituitarypituitary is controlled by hormones called hypothalamic releasing hormones and inhibitory hormones conducted to the anterior pituitary through hypothalamichypothalamic -- hypophysialhypophysial portalportal vesselsvessels .. • PosteriorPosterior pituitarypituitary secrets two hormones, which are synthesized within cell