Frameshift Variant in MFSD12 Explains the Mushroom Coat Color Dilution in Shetland Ponies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Use of Genomic Tools to Discover the Cause of Champagne Dilution Coat Color in Horses and to Map the Genetic Cause of Extreme Lordosis in American Saddlebred Horses
University of Kentucky UKnowledge Theses and Dissertations--Veterinary Science Veterinary Science 2014 USE OF GENOMIC TOOLS TO DISCOVER THE CAUSE OF CHAMPAGNE DILUTION COAT COLOR IN HORSES AND TO MAP THE GENETIC CAUSE OF EXTREME LORDOSIS IN AMERICAN SADDLEBRED HORSES Deborah G. Cook University of Kentucky, [email protected] Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Cook, Deborah G., "USE OF GENOMIC TOOLS TO DISCOVER THE CAUSE OF CHAMPAGNE DILUTION COAT COLOR IN HORSES AND TO MAP THE GENETIC CAUSE OF EXTREME LORDOSIS IN AMERICAN SADDLEBRED HORSES" (2014). Theses and Dissertations--Veterinary Science. 15. https://uknowledge.uky.edu/gluck_etds/15 This Doctoral Dissertation is brought to you for free and open access by the Veterinary Science at UKnowledge. It has been accepted for inclusion in Theses and Dissertations--Veterinary Science by an authorized administrator of UKnowledge. For more information, please contact [email protected]. STUDENT AGREEMENT: I represent that my thesis or dissertation and abstract are my original work. Proper attribution has been given to all outside sources. I understand that I am solely responsible for obtaining any needed copyright permissions. I have obtained needed written permission statement(s) from the owner(s) of each third-party copyrighted matter to be included in my work, allowing electronic distribution (if such use is not permitted by the fair use doctrine) which will be submitted to UKnowledge as Additional File. I hereby grant to The University of Kentucky and its agents the irrevocable, non-exclusive, and royalty-free license to archive and make accessible my work in whole or in part in all forms of media, now or hereafter known. -

Special Issue Horse Genetics DIRECTOR’S Message
CENTER FOR EQUINE HEALTH SCHOOL OF VETERINARY MEDICINE • UNIVERSITY OF CALIFORNIA, DAVIS SUMMER 2020 Special Issue Horse Genetics DIRECTOR’S Message s an equine genetics researcher, I am particularly excited to share A this special issue of the Horse Report with you. Inside, you will find a roadmap to many of the currently available equine genetic tests, including the AQHA “five-panel” test, and more. The equine genome sequence was published in 2009, the result of a years- long collaborative effort by the international equine research community. This resource drastically changed how researchers approach equine genetics and accelerated the rate of discovery. Increased availability and affordability allowed the application of advanced molecular tools to equine diseases and traits. As a result, genetic tests are available in a variety of breeds. Most available tests are for simple, Mendelian diseases and traits – those caused by a single gene or locus. Complex diseases and traits likely involve more than one gene and may be influenced by environmental effects. The 2018 release of a new equine genome sequence assembly, coupled with cost reductions that make whole-genome sequencing possible for large numbers of horses, are enabling research in these areas. As an equine geneticist and veterinarian, I am especially interested in applying whole genome sequencing and advanced diagnostic tools to equine precision medicine. This highly individualized approach will focus on early detection and prevention of disease, taking into account both genetic information and environmental factors. The idea is to target individuals based on their clinical condition as well as their unique body chemistry and genetics. -
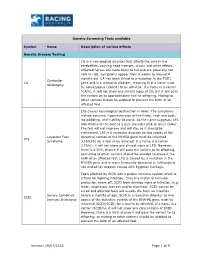
19/07/2018 Page 1 of 9 Genetic Screening Tests Available Symbol
Genetic Screening Tests available Symbol Name Description of variant effects Genetic Disease Testing CA is a neurological disorder that affects the cells in the cerebellum, causing head tremors, ataxia and other effects. Affected horses are more likely to fall and are generally not safe to ride. Symptoms appear from 6 weeks to around 4 months old. CA has been linked to a mutation in the TOE1 Cerebellar CA gene and is a recessive disorder, meaning that a horse must Abiotrophy be homozygous (CA/CA) to be affected. If a horse is a carrier (CA/n), it will not show any clinical signs of CA, but it will pass the variant on to approximately half its offspring. Mating to other carriers should be avoided to prevent the birth of an affected foal. LFS causes neurological dysfunction in foals. The symptoms include seizures, hyperextension of the limbs, neck and back, leg paddling, and inability to stand. As the name suggests, LFS also dilutes to the coat to a pale lavender pink or silver colour. The foal will not improve and will die, so it should be euthanised. LFS is a recessive disorder so two copies of the Lavender Foal defective version of the MYO5A gene must be inherited LFS Syndrome (LFS/LFS) for a foal to be affected. If a horse is a carrier (LFS/n), it will not show any clinical signs of LFS. However, there is a 50% chance it will pass the variant to its offspring, so mating to other carriers should be avoided to prevent the birth of an affected foal. -

The Base Colors: Black and Chestnut the Tail, Called “Foal Fringes.”The Lower Legs Can Be So Pale That It Is Let’S Begin with the Base Colors
Foal Color 4.08 3/20/08 2:18 PM Page 44 he safe arrival of a newborn foal is cause for celebration. months the sun bleaches the foal’s birth coat, altering its appear- After checking to make sure all is well with the mare and ance even more. Other environmental issues, such as type and her new addition, the questions start to fly. What gender quality of feed, also can have a profound effect on color. And as we is it? Which traits did the foal get from each parent? And shall see, some colors do change drastically in appearance with Twhat color is it, anyway? Many times this question is not easily age, such as gray and the roany type of sabino. Finally, when the answered unless the breeder has seen many foals, of many colors, foal shed occurs, the new color coming in often looks dramatical- throughout many foaling seasons. In the landmark 1939 movie, ly dark. Is it any wonder that so many foals are registered an incor- “The Wizard of Oz,” MGM used gelatin to dye the “Horse of a rect—and sometimes genetically impossible—color each year? Different Color,” but Mother Nature does a darn good job of cre- So how do you identify your foal’s color? First, let’s keep some ating the same spectacular special effects on her foals! basic rules of genetics in mind. Two chestnuts will only produce The foal’s color from birth to the foal shed (which generally chestnut; horses of the cream, dun, and silver dilutions must have occurs between three and four months of age) can change due to had at least one parent with that particular dilution themselves; many factors, prompting some breeders to describe their foal as and grays must always have one gray parent. -

Basic Horse Genetics
ALABAMA A&M AND AUBURN UNIVERSITIES Basic Horse Genetics ANR-1420 nderstanding the basic principles of genetics and Ugene-selection methods is essential for people in the horse-breeding business and is also beneficial to any horse owner when it comes to making decisions about a horse purchase, suitability, and utilization. Before getting into the basics of horse-breeding deci- sions, however, it is important that breeders under- stand the following terms. Chromosome - a rod-like body found in the cell nucleus that contains the genes. Chromosomes occur in pairs in all cells, with the exception of the sex cells (sperm and egg). Horses have 32 pairs of chromo- somes, and donkeys have 31 pairs. Gene - a small segment of chromosome (DNA) that contains the genetic code. Genes occur in pairs, one Quantitative traits - traits that show a continuous on each chromosome of a pair. range of phenotypic variation. Quantitative traits Alleles - the alternative states of a particular gene. The usually are controlled by more than one gene pair gene located at a fixed position on a chromosome will and are heavily influenced by environmental factors, contain a particular gene or one of its alleles. Multiple such as track condition, trainer expertise, and nutrition. alleles are possible. Because of these conditions, quantitative traits cannot be classified into distinct categories. Often, the impor- Genotype - the genetic makeup of an individual. With tant economic traits of livestock are quantitative—for alleles A and a, three possible genotypes are AA, Aa, example, cannon circumference and racing speed. and aa. Not all of these pairs of alleles will result in the same phenotype because pairs may have different Heritability - the portion of the total phenotypic modes of action. -

Impact of White Spotting Alleles, Including W20, on Phenotype in the American Paint Horse
bioRxiv preprint doi: https://doi.org/10.1101/678052; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Running Head: White spotting in the American Paint Horse Title: Impact of white spotting alleles, including W20, on phenotype in the American Paint Horse Samantha A. Brooks*, Katelyn M. Palermo*, Alisha Kahn*, and Jessica Hein# *University of Florida Department of Animal Sciences, UF Genetics Institute, Gainesville FL, 32611-0910 #American Paint Horse Association, Fort Worth TX, 76161-0023 Acknowledgments: The authors would like to thank the many APHA staff members for their efforts in submitting and collating the data analyzed in this study. Thanks to the UF undergraduate researchers who generously volunteered for data-entry work on this project: Hannah Hillard, Kalisse Horne, Rachel Kullman, Erica Riano, Matt Winter, Courtney McCreary, Rachel Shepherd, Anna Moskovitz, and Kaycie Miller. Our gratitude to Dr. Ernie Bailey for proofreading the manuscript. bioRxiv preprint doi: https://doi.org/10.1101/678052; this version posted June 21, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract: The American Paint Horse Association (APHA) officially records pedigree and performance information for their breed; these registered stock-type horses are valued for utility in work on the farm and ranch and as pleasure horses. As the name of the breed implies, the breed is also valued for attractive white spotting patterns on the coat. -

Appaloosa Coat Pattern - Leopard Print and Congenital Stationary Night Blindness (CSNB)
Appaloosa Coat Pattern - Leopard Print and Congenital Stationary Night Blindness (CSNB) Description: The coat patterns in spotted horses are genetically-related and are referred to as leopard spotting complex. The causal mutation associated with leopard complex are also linked to abnormalities in the eyes and vision (CSNB). These patterns are most closely identified with the Appaloosa horse breed, though its presence in breeds from Asia (Tiger Horses) to western Europe (Knabstrupper or Knabstrup) has indicated that it is due to a very ancient mutation. European cave paintings have recorded spotted horses indicating that the phenotype has existed for 20,000 years prior to their domestication, some 5,000 years ago. The Appaloosa horse descended from horses brought to American by the Spanish in the 16th century. The name Appaloosa comes from the Palouse river that ran through the Nez Percé Indians territory. The breed is best characterized by its spotted coat pattern commonly referred to as "leopard-complex". The leopard-spotted coat pattern has also been documented in several other horse breeds, including the Pony of the Americas, the Nez Percé Horse and several gaited horse breeds. Within the Appaloosa breed there is a wide range of body types, stemming from the influence of multiple breeds of horses including Arabian blood lines which were introduced during the late 19th century. Although commonly recognized by their colorful coat patterns, Appaloosa horses have three additional identifiable characteristics including mottled skin around the nose, lips, and genitals, striped hooves and white sclera round the eyes. In 2003, researchers linked the positional candidate gene for leopard complex (LP) to the TRPM1 gene on chromosome 1. -

OUR HOUSE DESIGNS ACTIVE LEATHER / FABRIC LIST - OCTOBER MARKET 2018 Listed Below Are All Leathers That Are Active
OUR HOUSE DESIGNS ACTIVE LEATHER / FABRIC LIST - OCTOBER MARKET 2018 Listed below are all leathers that are active. Please remove swatches that are not on this list. * REGULAR LEATHERS * Leather # Color Name Leather # Color Name Leather # Color Name 302A-51 Cherry 382A-71 Teal 404P-85 Tigers Eye 305A-81 Brown 383A-21 Citron 404P-86 Fudgesicle 333N-11 Storm Cloud 388A-81 Espresso 404P-87 Nutmeg 349A-71 Peacock 390N-81 Longhorn 406A-11 Ash 350A-71 Bell Pepper 398A-21 Sand 410A-11 Stone 351A-11 Ivory 398A-62 Black 412A-81 Dark Rum 351A-61 Crystal 398A-71 Hazy 416A-11 Mist 356A-81 Mulch 399P-81 Coconut Husk 416A-31 Heaven 357A-81 Terra 400P-11 Moon Dust 417A-31 Spa 359AC-21 Camel 400P-61 Asphalt 424A-61 Black 359AC-31 Sailor 400P-62 Ashe 425A-11 Hand Wipe 359AC-51 Cherry Red 400P-82 Umber 466A-82 Saddle 359AC-52 Brick 401A-31 Sapphire 490A-83 Wolf 359AC-61 Midnight 401A-71 Evergreen 494P-11 Opalite 359AC-62 Smoke 402N-31 Sailboat 494P-13 Sandstone 359AC-71 Fern 402N-61 Slate 494P-61 Flagstone 359AC-72 Aqua 402N-81 Molasses 494P-62 Agate 359AC-81 Tundra 403N-31 Dark Blue 494P-81 Bronzite 359AC-82 Milk Chocolate 404P-11 Shale 494P-82 Tiger Eye 359AC-83 Tile 404P-12 Cobblestone 494P-83 Marble 359AC-84 Hazy 404P-13 Malt 495P-11 Mink 359AC-85 Espresso 404P-14 Chantillie 495P-61 Lynx 369A-11 Hand Glazed 404P-21 Cider 522A-81 Brown 371P-11 Shadow 404P-31 Azure 524A-61 Hazy Black 371P-81 Mud Cakes 404P-32 Oceanic 531A-61 Fog 373P-11 Starlight 404P-52 Red Velvet 531A-81 Cashew 373P-12 Fog 404P-53 Bitters 531A-83 Espresso 378A-81 Pecan 404P-61 Heather -

4COLOURS of the FJORD HORSE2014
COLOURS of the FJORD HORSE By Tor Nestaas. The colours of the Fjord Horse are a variety of dun colours. The central –Asian Wild Horse, the Prezewalski horse and the European Wild Horse, the Tarpan have the same types of colour and these colours are seen to be the original colours of the wild horse. The colour is also called a primitive colour or “viltfarge“. (Protective colour ). The pure dun colours are brown dun, red dun and grey dun but variations such as “uls dun “ and yellow dun are also seen . These five colours are recognized as pure fjord horse colours. Earlier these colours could have different names in different districts, but in 1922 the Department of Agriculture decided on the names that are used today. The Annual General Meeting of NFHL in 1980 agreed that these five colours are all typical fjord horse colours and should be treated equally. Colour variations The brown dun colour (brunblakk ) is dominant, 85-90 % of all fjord horses have this colour which can be of lighter or darker shades.The body colour is pale yellow brown and can vary from cream yellow to almost light brown. The “midtstøl “the middle darker stripe through the mane, carrying on as the dorsal stripe* to the snow -shute* ( all considered as primitive markings ) is black or dark brown. In paler individuals the forelock and “sides“ (of the upright mane ) are white but darker in darker individuals. * Sometimes called eel or list. Fanshaped growth of hairs at the top of the tail Ulsdun (ulsblakk ) is a variation of brown dun because of a factor which reduces the production of pigment ; so called diluted colour. -

Features of Coat Color and Markings and Impact of Dun Factor on Vyatka Horse Breed
BIO Web of Conferences 17, 00202 (2020) https://doi.org/10.1051/bioconf/20201700202 FIES 2019 Features of coat color and markings and impact of dun factor on Vyatka horse breed Natalia F. Belousova1, Svetlana P. Bass2, Svetlana A. Zinoveva3, Sergei A. Kozlov3,*, and Sergei S. Markin3 1All-Russian Research Institute of Horse Breeding (VNIIK), 391105 Divovo Village, Ryazan Region, Russia 2Izhevsk State Agricultural Academy, 426069 Izhevsk, Russia 3Moscow State Academy of Veterinary Medicine and Biotechnology – MVA named after K.I. Skryabin, 109472 Moscow, Russia Abstract. The predominant coat colors in Vyatka horse breed are bay-brown (69.6 %) and mousey (20.8 %). Among the genotyped livestock, three genotypes of the base bay coat color (EE/AA, EE/Aa, Ee/AA) and two genotypes of the base solid blackcock (EE/a/a, Ee/aa) have been detected. The proportion of horses with Cr allele is 2.1 %. In Vyatka horse breed, three isabelline-brown horses (Cr/Cr) have been recorded and the presence of W20n allele was detected. Among the horses genotyped, 35.5 % are DD homozygous, 61.3 % are heterozygous (Dd1, Dd2), 3.2 % have the nd2/nd2 genotype. Allele d2 against the background of D does not always cause the presence of “wild” markings, unlike D/D. The influence of Dun-factor on the depigmentation area has not been detected. 39.9 % of horses have white markings (including 30 % of stallions), which are mainly facial markings (59.8 %), less often they are leg markings (21.6 %) or both facial and leg markings (18, 6 %). 1 Introduction At the same time, the chestnut (red and brown) coat color has become rather rare in the breed. -

Distinguishable Markings Guide Color Chart
Gypsy Horse Registry of America, Inc. Color & Pattern Guide Distinguishable Markings Guide Star Star Star, Strip Blaze Bald Apron & Strip Snip & Snip Front Front Front Front Rear Rear Rear Rear Coronet Fetlock Sock Stocking Coronet Fetlock Sock Stocking Color Chart Light yellow to reddish brown body to dark grey with dorsal Bay Chestnut body color with black mane, tail & points Dun stripe, may have primitive markings on head, withers and legs. Black Black hairs, no brown, red or tan hairs visible. Grey Born any dark color, gradually turning white with age Smokey to mouse grey (not a roan) with darker head with black A very dark brown, almost black coat with lighter brown Brown Grullo mane, tail & points, may have dorsal stripe & primitive markings highlights on muzzle, flanks & inside legs. on head, withers and legs. Buckskin Light tan to golden body with black mane, tail & points Palomino Clear yellow to rich golden coat with white mane and tail Silver Dark steel grey, chocolate to light sepia brown, light to heavy Chestnut Red with red or flaxen mane & tail Dapple dappling with flaxen to white mane, tail & points Color Pattern Guide TOBIANO (toe-bee-ah-no): SABINO (sah-bee-no): SOLID The most common type of white spotting seen This is a common pattern also known as Body is a solid color, black, chestnut, bay, buckskin, on horses. The white areas usually have a flecked roan, roan or Blagdon in Europe. silver dapple, palomino, etc. May have white on face distinct, sharp edge to them. The back is Blue eyes are common, head and legs (star, blaze, snip or strip, etc.) and /or lower legs usually crossed at some point by white. -

Genetics and Genomics of Mammalian Pigment Patterns
GENETICS AND GENOMICS OF MAMMALIAN PIGMENT PATTERNS A DISSERTATION SUBMITTED TO THE DEPARTMENT OF GENETICS AND THE COMMITTEE ON GRADUATE STUDIES OF STANFORD UNIVERSITY IN PARTIAL FUFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY Lewis Zuocheng Hong August 2011 © 2011 by Zuocheng Lewis Hong. All Rights Reserved. Re-distributed by Stanford University under license with the author. This work is licensed under a Creative Commons Attribution- Noncommercial 3.0 United States License. http://creativecommons.org/licenses/by-nc/3.0/us/ This dissertation is online at: http://purl.stanford.edu/jx191nt1141 ii I certify that I have read this dissertation and that, in my opinion, it is fully adequate in scope and quality as a dissertation for the degree of Doctor of Philosophy. Gregory Barsh, Primary Adviser I certify that I have read this dissertation and that, in my opinion, it is fully adequate in scope and quality as a dissertation for the degree of Doctor of Philosophy. Andrew Fire I certify that I have read this dissertation and that, in my opinion, it is fully adequate in scope and quality as a dissertation for the degree of Doctor of Philosophy. David Kingsley I certify that I have read this dissertation and that, in my opinion, it is fully adequate in scope and quality as a dissertation for the degree of Doctor of Philosophy. Arend Sidow Approved for the Stanford University Committee on Graduate Studies. Patricia J. Gumport, Vice Provost Graduate Education This signature page was generated electronically upon submission of this dissertation in electronic format. An original signed hard copy of the signature page is on file in University Archives.