Revision of Freshwater Shrimps Belonging to Caridina Weberi Complex
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Two Freshwater Shrimp Species of the Genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the Description of a New Species
A peer-reviewed open-access journal ZooKeys 923: 15–32 (2020) Caridina tetrazona 15 doi: 10.3897/zookeys.923.48593 RESEarcH articLE http://zookeys.pensoft.net Launched to accelerate biodiversity research Two freshwater shrimp species of the genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the description of a new species Qing-Hua Chen1, Wen-Jian Chen2, Xiao-Zhuang Zheng2, Zhao-Liang Guo2 1 South China Institute of Environmental Sciences, Ministry of Ecology and Environment, Guangzhou 510520, Guangdong Province, China 2 Department of Animal Science, School of Life Science and Enginee- ring, Foshan University, Foshan 528231, Guangdong Province, China Corresponding author: Zhao-Liang Guo ([email protected]) Academic editor: I.S. Wehrtmann | Received 19 November 2019 | Accepted 7 February 2020 | Published 1 April 2020 http://zoobank.org/138A88CC-DF41-437A-BA1A-CB93E3E36D62 Citation: Chen Q-H, Chen W-J, Zheng X-Z, Guo Z-L (2020) Two freshwater shrimp species of the genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the description of a new species. ZooKeys 923: 15–32. https://doi.org/10.3897/zookeys.923.48593 Abstract A faunistic and ecological survey was conducted to document the diversity of freshwater atyid shrimps of Dawanshan Island. Two species of Caridina that occur on this island were documented and discussed. One of these, Caridina tetrazona sp. nov. is described and illustrated as new to science. It can be easily distinguished from its congeners based on a combination of characters, which includes a short rostrum, the shape of the endopod of the male first pleopod, the segmental ratios of antennular peduncle and third maxilliped, the slender scaphocerite, and the absence of a median projection on the posterior margin. -

A Genetical and Ecological Diversity of Fresh Water Prawns Macrobrachium Canarae and Caridina Gracilirostris from Kanyakumari Dist., Tamil Nadu, India
International Journal of Genetic Engineering and Biotechnology. ISSN 0974-3073 Volume 2, Number 1 (2011), pp. 23-32 © International Research Publication House http://www.irphouse.com A Genetical and Ecological Diversity of Fresh Water Prawns Macrobrachium Canarae and Caridina Gracilirostris from Kanyakumari Dist., Tamil Nadu, India Siva Ranjanee S.1 and Mariapan N.2 1Department of Biotechnology, Vels University, Pallavaram, Chennai, India 2Senior Medical Writer, SIRO Clinpharm Pvt. Ltd., Thane (W) 400 607. India E-mail: [email protected], [email protected] Abstract Fresh water prawns are cultured widely around the world but little is known about the levels and patterns of genetic diversity. This paper reports the RAPD analysis of two species of fresh water prawns of Atydiae and Palaemonidiae family and the genus Macrobrachium and Caridina collected from Kanyakumari district. Specimens are identified using species-specific morphological characteristics. Morphological characters have limitations of describing intra specific genetic diversity as they are polygenic and expressions can be modified by the environment. The advent of molecular technique made possible not only the genetic analysis and also the study of evolutionary relationship. Molecular analysis of the morphologically identified species are done by extracting the DNA from the animal and concentration of DNA is measured using Nanodrop spectrophotometer at a wavelength of 280nm. Further analysis was performed with RAPD (Rapid Amplified Polymorphic DNA) a PCR (Polymerase Chain Reaction) based technique. It consist of genomic DNA, amplified with randomly constructed oligonucleotides. Specific quantity of extracted DNA was then amplified by multiplex PCR using random primers and 16S rRNA gene products was then analyzed using a bioanalyzer. -

Note Food and Feeding Habits of Macrobrachium
Indian J. Fish., 60(4) : 131-135, 2013 Note Food and feeding habits of Macrobrachium lar (Decapoda, Palaemonidae) from Andaman and Nicobar Islands, India S. N. SETHI1, NAGESH RAM2 AND V. VENKATESAN3 1Chennai Research Centre of Central Marine Fisheries Research Institute, 75 – Santhome High Road R. A. Puram, Chennai-600 028,Tamil Nadu, India 2Central Agricultural Research Institute (CARI), Port Blair-744 103, Andaman and Nicobar Islands, India 3Central Marine Fisheries Research Institute, Cochin -682 018, Kerala, India e-mail: [email protected] ABSTRACT The stomach content of the freshwater prawn Macrobrachium lar from the inland water bodies of Andaman and Nicobar Islands were analysed in relation to size, sex and season during the period from January to December 2008. Feeding habits of M. lar were studied using the index of preponderance and feeding intensity methods. Mainly organic detritus, supplemented by animal and plant materials formed the food in all stages of M. lar. The animal food items mainly consisted of aquatic insects, polychaetes, other crustaceans, fish, molluscs and zooplankton. The plant matter was chiefly composed of fragments of aquatic plants, planktonic algae and diatoms. Variation in diet in relation to size, sex or season were found insignificant. Though, all size groups of this species feed animal and plant materials, it exhibited slight preference for animal food especially in larger size groups. Females appear to feed more actively than the males. Feeding intensity was more in monsoon season compared to dry season. Results indicate that the adults are mostly predators of slow moving benthic invertebrates rather than detritus feeders/scavengers. -

Species-Edition-Melanesian-Geo.Pdf
Nature Melanesian www.melanesiangeo.com Geo Tranquility 6 14 18 24 34 66 72 74 82 6 Herping the final frontier 42 Seahabitats and dugongs in the Lau Lagoon 10 Community-based response to protecting biodiversity in East 46 Herping the sunset islands Kwaio, Solomon Islands 50 Freshwater secrets Ocean 14 Leatherback turtle community monitoring 54 Freshwater hidden treasures 18 Monkey-faced bats and flying foxes 58 Choiseul Island: A biogeographic in the Western Solomon Islands stepping-stone for reptiles and amphibians of the Solomon Islands 22 The diversity and resilience of flying foxes to logging 64 Conservation Development 24 Feasibility studies for conserving 66 Chasing clouds Santa Cruz Ground-dove 72 Tetepare’s turtle rodeo and their 26 Network Building: Building a conservation effort network to meet local and national development aspirations in 74 Secrets of Tetepare Culture Western Province 76 Understanding plant & kastom 28 Local rangers undergo legal knowledge on Tetepare training 78 Grassroots approach to Marine 30 Propagation techniques for Tubi Management 34 Phantoms of the forest 82 Conservation in Solomon Islands: acts without actions 38 Choiseul Island: Protecting Mt Cover page The newly discovered Vangunu Maetambe to Kolombangara River Island endemic rat, Uromys vika. Image watershed credit: Velizar Simeonovski, Field Museum. wildernesssolomons.com WWW.MELANESIANGEO.COM | 3 Melanesian EDITORS NOTE Geo PRODUCTION TEAM Government Of Founder/Editor: Patrick Pikacha of the priority species listed in the Critical Ecosystem [email protected] Solomon Islands Hails Partnership Fund’s investment strategy for the East Assistant editor: Tamara Osborne Melanesian Islands. [email protected] Barana Community The Critical Ecosystem Partnership Fund (CEPF) Contributing editor: David Boseto [email protected] is designed to safeguard Earth’s most biologically rich Prepress layout: Patrick Pikacha Nature Park Initiative and threatened regions, known as biodiversity hotspots. -

Caridina Variabilirostris (Crustacea: Decapoda: Atyidae), a New Species
Caridina variabilirostris (Crustacea: Decapoda: Atyidae), a new species of freshwater shrimp from Pohnpei (Micronesia) Valentin de Mazancourt, Gerard Marquet, Philippe Keith To cite this version: Valentin de Mazancourt, Gerard Marquet, Philippe Keith. Caridina variabilirostris (Crustacea: De- capoda: Atyidae), a new species of freshwater shrimp from Pohnpei (Micronesia). European Journal of Taxonomy, Consortium of European Natural History Museums, 2018, pp.1-16. 10.5852/ejt.2018.453. hal-02171169 HAL Id: hal-02171169 https://hal.archives-ouvertes.fr/hal-02171169 Submitted on 2 Jul 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. European Journal of Taxonomy 453: 1–16 ISSN 2118-9773 https://doi.org/10.5852/ejt.2018.453 www.europeanjournaloftaxonomy.eu 2018 · Mazancourt V. de et al. This work is licensed under a Creative Commons Attribution 3.0 License. Research article urn:lsid:zoobank.org:pub:445C8252-5FA2-49AA-AB89-E1C2DDEBEE7F Caridina variabilirostris (Crustacea: Decapoda: Atyidae), a new species of freshwater shrimp from Pohnpei (Micronesia) Valentin de MAZANCOURT 1,*, Gerard MARQUET 2 & Philippe KEITH 3 1,2,3 Muséum national d’Histoire naturelle, Département Adaptations du Vivant, UMR 7208, CP026, 57, rue Cuvier, 75231 Paris, Cedex 05, France. -

Caridina Wild Type Final 1
FRESHWATER SHRIMP WILD TYPE BY CHRIS LUKHAUP Paracaridina meridionalis During the course of evolution, Caridina logemanni They are also the most widely spread Paracaridina meridionalis freshwater dwarf shrimp have spread shrimp in the aquarium hobby. from the estuaries of larger rivers to However, recent research has found rapidly-flowing creeks in the that this genus is in urgent need of mountains,to enormous lakes, the a scientific review and re-structuring smallest trickles of water and even to as there are many discrepancies to cave waters. They are found from be found. The genus Neocaridina tropic to temperate climates today, has up until now been represented however, they have not conquered by 30 species, and has also found wide distribution in the hobby. Paracaridina meridionalis the arctic regions yet. The abundance Paracaridina meridionalis of different habitats has resulted in a Within the Genus Paracaridina just great variability in shrimp species and very few species have been described in stunning forms. Their sometimes but nevertheless some of them are truly impressive colours and patterns found in the shrimpkeeping hobby are the result of their adaptation to the different living conditions in their habitats. Enjoy and keep on shrimping ! With over 290 species, shrimp of the Paracaridina meridionalis genus Caridina are one of the most Paracaridina meridionalis diverse groups within the Atyidae family. Paracaridina meridionalis Paracaridina meridionalis Paracaridina meridionalis Paracaridina meridionalis Paracaridina meridionalis Caridina logemanni Caridina sp. “Vietnam” Caridina serrata Caridina serrata Caridina serrata Caridina trifasciata Caridina trifasciata Caridina trifasciata Caridina trifasciata Caridina sumatrensis paraCaridina sp. Caridina breviata Atyaephyra desmaresti Caridina simoni Caridina gracilirostris caridina serratirostris caridina serratirostris caridina serratirostris caridina serratirostris caridina sp” .Vietnam Blue” Caridina sp. -

Caridina Propinqua) Ecological Risk Screening Summary
Bengal Caridina (Caridina propinqua) Ecological Risk Screening Summary U.S. Fish & Wildlife Service, July 2017 Revised, August 2017 Web Version, 11/17/2017 1 Native Range and Status in the United States Native Range From De Grave and Cai (2013): “The species is widespread from Bangladesh and India through to the Ryukyus and southwards to Singapore (Cai and Shokita 2006).” “Bangladesh; India (Orissa); Japan; Malaysia (Peninsular Malaysia); Philippines; Singapore; Sri Lanka; Thailand” Status in the United States This species has not been reported as introduced or established in the United States. This species is in trade in the U.S. From Bob’s Tropical Plants (2017): “Caridina cf. propinqua orange shrimp […] $2.75 tax excl.” “They are still relatively rare in the hobby, but are not difficult to maintain.” 1 Means of Introductions in the United States This species has not been reported as introduced or established in the United States. Remarks From GBIF (2016): “SYNONYMS Caridina blancoi Chace, 1997 Caridina hainanensis Liang & Yan, 1983” 2 Biology and Ecology Taxonomic Hierarchy and Taxonomic Standing From ITIS (2017): “Kingdom Animalia Subkingdom Bilateria Infrakingdom Protostomia Superphylum Ecdysozoa Phylum Arthropoda Subphylum Crustacea Class Malacostraca Subclass Eumalacostraca Superorder Eucarida Order Decapoda Suborder Pleocyemata Infraorder Caridea Superfamily Atyoidea Family Atyidae Genus Caridina Species Caridina propinqua De Man, 1908 – bengal caridina” “Taxonomic Status: Current Standing: valid” Size, Weight, and Age Range From Cai and Shokita (2006): “Material examined […] 11 males, cl [carapace length] 2.6–3.3 mm, 6 females, cl 3.5–3.8 mm, 16 ovigerous females, cl 3.4–4.0 mm […]” 2 Environment From Cai and Shokita (2006): “Lower reaches of rivers or mountain streams which discharge to the sea. -
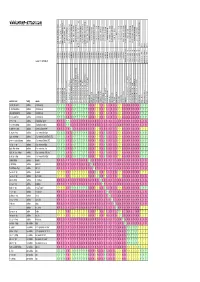
Crossbreeding/Interbreeding Chart Gotten from Web-Site
www.shrimp-attack.com nn blue blue pearl rib bee line ninja shrimp ninja green shrimp orchid shrimp yellow shrimp yellow sakura shrimp tuepfel shrimp orange shrimp yamato shrimp cardinal shrimp red tiger shrimp new new bee shrimp new bee shrimp new new bee shrimp new bee shrimp snowball shrimp snowball goldflake shrimp harlequin shrimp harlequin shrimp blue blue tiger shrimp red cherry shrimp red shrimp hawaii black tiger shrimp crystal red shrimp panda taiwan Bee bumble beebumble shrimp indian zebraindian shrimp crystal black shrimp blue blue bolt taiwan bee red ruby Taiwan bee Towuti/Matano Tiger king king kong Taiwan bee marmoreal dwarfshrimp crystal white bee shrimp golden golden chrystal red shrimp spec. spec. spec. cardina cardina cardina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina caridina neocaridina neocaridina neocaridina neocaridina neocaridina neocaridina version 1.1. / 2010-04-20 halocaridina paracaridina paracaridina paracaridina common name family species cf.cf. cantonensiscantonensis 'Golden 'Golden CRS' CRS' cf.cf. cantonensiscantonensis redred tiger tiger cf.cf. cantonensiscantonensis sp.sp. 'bee' 'bee' cf.cf. cantonensiscantonensis sp.sp. 'white'white bee' bee' cf.cf. cantonensiscantonensis blackblack tiger tiger dennerli glaubrechti holthuisi maculata meridionalis multidentata propinqua serrata 'tuepfel' serratirostris spinata spongicola striata tenuirostris tumida venusta woltereckae rubra cf.cf. var. var.zhangjiajiensis zhangjiajiensis 'blue' 'blue' cf.cf. var. var.zhangjiajiensis zhangjiajiensis 'white' 'white' heteropodaheteropoda var.var. 'red' 'red' heteropodaheteropoda var.var. 'sakura' 'sakura' heteropodaheteropoda var.var. 'yellow' 'yellow' palmata blue bee princess bee larry shrimp cantonensis sp. cantonensis sp. -

De Grave & Fransen. Carideorum Catalogus
De Grave & Fransen. Carideorum catalogus (Crustacea: Decapoda). Zool. Med. Leiden 85 (2011) 407 Fig. 48. Synalpheus hemphilli Coutière, 1909. Photo by Arthur Anker. Synalpheus iphinoe De Man, 1909a = Synalpheus Iphinoë De Man, 1909a: 116. [8°23'.5S 119°4'.6E, Sapeh-strait, 70 m; Madura-bay and other localities in the southern part of Molo-strait, 54-90 m; Banda-anchorage, 9-36 m; Rumah-ku- da-bay, Roma-island, 36 m] Synalpheus iocasta De Man, 1909a = Synalpheus Iocasta De Man, 1909a: 119. [Makassar and surroundings, up to 32 m; 0°58'.5N 122°42'.5E, west of Kwadang-bay-entrance, 72 m; Anchorage north of Salomakiëe (Damar) is- land, 45 m; 1°42'.5S 130°47'.5E, 32 m; 4°20'S 122°58'E, between islands of Wowoni and Buton, northern entrance of Buton-strait, 75-94 m; Banda-anchorage, 9-36 m; Anchorage off Pulu Jedan, east coast of Aru-islands (Pearl-banks), 13 m; 5°28'.2S 134°53'.9E, 57 m; 8°25'.2S 127°18'.4E, an- chorage between Nusa Besi and the N.E. point of Timor, 27-54 m; 8°39'.1 127°4'.4E, anchorage south coast of Timor, 34 m; Mid-channel in Solor-strait off Kampong Menanga, 113 m; 8°30'S 119°7'.5E, 73 m] Synalpheus irie MacDonald, Hultgren & Duffy, 2009: 25; Figs 11-16; Plate 3C-D. [fore-reef (near M1 chan- nel marker), 18°28.083'N 77°23.289'W, from canals of Auletta cf. sycinularia] Synalpheus jedanensis De Man, 1909a: 117. [Anchorage off Pulu Jedan, east coast of Aru-islands (Pearl- banks), 13 m] Synalpheus kensleyi (Ríos & Duffy, 2007) = Zuzalpheus kensleyi Ríos & Duffy, 2007: 41; Figs 18-22; Plate 3. -

Assessment of the Risk to Norwegian Biodiversity from Import and Keeping of Crustaceans in Freshwater Aquaria
VKM Report 2021: 02 Assessment of the risk to Norwegian biodiversity from import and keeping of crustaceans in freshwater aquaria Scientific Opinion of the Panel on Alien Organisms and Trade in Endangered Species of the Norwegian Scientific Committee for Food and Environment VKM Report 2021: 02 Assessment of the risk to Norwegian biodiversity from import and keeping of crustaceans in freshwater aquaria. Scientific Opinion of the Panel on Alien Organisms and trade in Endangered Species (CITES) of the Norwegian Scientific Committee for Food and Environment 15.02.2021 ISBN: 978-82-8259-356-4 ISSN: 2535-4019 Norwegian Scientific Committee for Food and Environment (VKM) Postboks 222 Skøyen 0213 Oslo Norway Phone: +47 21 62 28 00 Email: [email protected] vkm.no vkm.no/english Cover photo: Mohammed Anwarul Kabir Choudhury/Mostphotos.com Suggested citation: VKM, Gaute Velle, Lennart Edsman, Charlotte Evangelista, Stein Ivar Johnsen, Martin Malmstrøm, Trude Vrålstad, Hugo de Boer, Katrine Eldegard, Kjetil Hindar, Lars Robert Hole, Johanna Järnegren, Kyrre Kausrud, Inger Måren, Erlend B. Nilsen, Eli Rueness, Eva B. Thorstad and Anders Nielsen (2021). Assessment of the risk to Norwegian biodiversity from import and keeping of crustaceans in freshwater aquaria. Scientific Opinion of the Panel on Alien Organisms and trade in Endangered Species (CITES) of the Norwegian Scientific Committee for Food and Environment. VKM report 2021:02, ISBN: 978-82-8259- 356-4, ISSN: 2535-4019. Norwegian Scientific Committee for Food and Environment (VKM), Oslo, Norway. 2 Assessment of the risk to Norwegian biodiversity from import and keeping of crustaceans in freshwater aquaria Preparation of the opinion The Norwegian Scientific Committee for Food and Environment (Vitenskapskomiteen for mat og miljø, VKM) appointed a project group to draft the opinion. -

Lines of Evidence for the Validity of Caridina Buehleri Roux (Crustacea : Decapoda : Atyidae) and for C
When molecules and morphology work together: lines of evidence for the validity of Caridina buehleri Roux (Crustacea : Decapoda : Atyidae) and for C. gueryi Marquet, Keith & Kalfatak as its junior synonym Valentin de Mazancourt, Gerard Marquet, Werner Klotz, Philippe Keith, Magalie Castelin To cite this version: Valentin de Mazancourt, Gerard Marquet, Werner Klotz, Philippe Keith, Magalie Castelin. When molecules and morphology work together: lines of evidence for the validity of Caridina buehleri Roux (Crustacea : Decapoda : Atyidae) and for C. gueryi Marquet, Keith & Kalfatak as its junior synonym. Invertebrate Systematics, CSIRO Publishing, 2017, 31 (2), pp.220. 10.1071/IS16044. hal-02171153 HAL Id: hal-02171153 https://hal.archives-ouvertes.fr/hal-02171153 Submitted on 2 Jul 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 1 When molecules and morphology work together: lines of evidence for the validity of 2 Caridina buehleri Roux, 1934 (Crustacea: Decapoda: Atyidae) and for C. gueryi Marquet, 3 Keith & Kalfatak, 2009 as its junior synonym. 4 VALENTIN de MAZANCOURT1*, GERARD MARQUET1, WERNER KLOTZ3, PHILIPPE 5 KEITH1 AND MAGALIE CASTELIN1-2 6 7 1 Muséum national d’Histoire naturelle, DMPA, UMR 7208, CP026, 57, rue Cuvier, F-75231 8 Paris, Cedex 05 9 2 Aquatic Animal Health Section, Fisheries and Oceans Canada, Pacific Biological Station, 10 3190 Hammond Bay Road, Nanaimo, BC, Canada V9T 6N7 11 3 Wiesenweg 1, A-6063 Rum, Austria. -
![[PDF] Freshwater Crustacea of the Mimika Region, New Guinea](https://docslib.b-cdn.net/cover/9537/pdf-freshwater-crustacea-of-the-mimika-region-new-guinea-1739537.webp)
[PDF] Freshwater Crustacea of the Mimika Region, New Guinea
BioAccess Australia Short, J. W., 2009. Freshwater Crustacea of the Mimika region – New Guinea. PT Freeport Indonesia: Kuala Kencana, ix + 96 pp. Other publications by John W. Short BioAccess Australia biodiversity consultancy and publishing Freshwater Crustacea of the Mimika Region New Guinea John W. Short Published by: PT Freeport Indonesia Enviromental Department Jl. Mandala Raya Selatan No. 1, Kuala Kencana, Timika 99920 Papua Design: John W. Short Cover Design: Hilman Ansori Printed by: PT. Indonesia Printer © John W. Short, 2009. All material in this book is copyright and may not be reproduced except with the written permission of the publishers. ISBN 978-979-97593-3-4 Cover illustration: Southern New Guinean River Prawn, Macrobrachium sp. 1 (Photo by John W. Short) Title Page Illustrations: Spiny Pacific Fan Shrimp, Atyopsis spinipes (after Cowles, 1914), top; Knob- fingered River Prawn, Macrobrachium mammillodactylus (Photo by John W. Short), bottom. Foreword As in my previous foreword for the Mangrove Estuary Crabs of the Mimika Region book, I herewith would like to thank Dr. John W. Short and all contributors in the Freshwater Crustacea of the Mimika Region book for their hard work and dedication towards documenting the freshwater crustacean habitat in the region for the benefit of broader society. This is the seventh biodiversity book published by PT Freeport Indonesia, in its quest to continuously disseminate knowledge related to biodiversity to the public. PT Freeport Indonesia, in more than 40 years of its operations in Papua, finds itself located in one of the most exotic and unique environments in the world. The island is one of the most biologically diverse mangrove estuary ecostystems and its 1.75 million hectares of mangrove vegetation is the largest in Indonesia.