Thermostable Designed Ankyrin Repeat Proteins (Darpins) As Building Blocks for Innovative Drugs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ankrd9 Is a Metabolically-Controlled Regulator of Impdh2 Abundance and Macro-Assembly
ANKRD9 IS A METABOLICALLY-CONTROLLED REGULATOR OF IMPDH2 ABUNDANCE AND MACRO-ASSEMBLY by Dawn Hayward A dissertation submitted to The Johns Hopkins University in conformity with the requirements of the degree of Doctor of Philosophy Baltimore, Maryland April 2019 ABSTRACT Members of a large family of Ankyrin Repeat Domains proteins (ANKRD) regulate numerous cellular processes by binding and changing properties of specific protein targets. We show that interactions with a target protein and the functional outcomes can be markedly altered by cells’ metabolic state. ANKRD9 facilitates degradation of inosine monophosphate dehydrogenase 2 (IMPDH2), the rate-limiting enzyme in GTP biosynthesis. Under basal conditions ANKRD9 is largely segregated from the cytosolic IMPDH2 by binding to vesicles. Upon nutrient limitation, ANKRD9 loses association with vesicles and assembles with IMPDH2 into rod-like structures, in which IMPDH2 is stable. Inhibition of IMPDH2 with Ribavirin favors ANKRD9 binding to rods. The IMPDH2/ANKRD9 assembly is reversed by guanosine, which restores association of ANKRD9 with vesicles. The conserved Cys109Cys110 motif in ANKRD9 is required for the vesicles-to-rods transition as well as binding and regulation of IMPDH2. ANKRD9 knockdown increases IMPDH2 levels and prevents formation of IMPDH2 rods upon nutrient limitation. Thus, the status of guanosine pools affects the mode of ANKRD9 action towards IMPDH2. Advisor: Dr. Svetlana Lutsenko, Department of Physiology, Johns Hopkins University School of Medicine Second reader: -

Repeat Proteins Challenge the Concept of Structural Domains
844 Biochemical Society Transactions (2015) Volume 43, part 5 Repeat proteins challenge the concept of structural domains Rocıo´ Espada*1, R. Gonzalo Parra*1, Manfred J. Sippl†, Thierry Mora‡, Aleksandra M. Walczak§ and Diego U. Ferreiro*2 *Protein Physiology Lab, Dep de Qu´ımica Biologica, ´ Facultad de Ciencias Exactas y Naturales, UBA-CONICET-IQUIBICEN, Buenos Aires, C1430EGA, Argentina †Center of Applied Molecular Engineering, Division of Bioinformatics, Department of Molecular Biology, University of Salzburg, 5020 Salzburg, Austria ‡Laboratoire de physique statistique, CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France §Laboratoire de physique theorique, ´ CNRS, UPMC and Ecole normale superieure, ´ 24 rue Lhomond, 75005 Paris, France Abstract Structural domains are believed to be modules within proteins that can fold and function independently. Some proteins show tandem repetitions of apparent modular structure that do not fold independently, but rather co-operate in stabilizing structural forms that comprise several repeat-units. For many natural repeat-proteins, it has been shown that weak energetic links between repeats lead to the breakdown of co-operativity and the appearance of folding sub-domains within an apparently regular repeat array. The quasi-1D architecture of repeat-proteins is crucial in detailing how the local energetic balances can modulate the folding dynamics of these proteins, which can be related to the physiological behaviour of these ubiquitous biological systems. Introduction and between repeats, challenging the concept of structural It was early on noted that many natural proteins typically domain. collapse stretches of amino acid chains into compact units, defining structural domains [1]. These domains typically correlate with biological activities and many modern proteins can be described as composed by novel ‘domain arrange- Definition of the repeat-units ments’ [2]. -

A Structural Guide to Proteins of the NF-Kb Signaling Module
Downloaded from http://cshperspectives.cshlp.org/ on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press A Structural Guide to Proteins of the NF-kB Signaling Module Tom Huxford1 and Gourisankar Ghosh2 1Department of Chemistry and Biochemistry, San Diego State University, 5500 Campanile Drive, San Diego, California 92182-1030 2Department of Chemistry and Biochemistry, University of California, San Diego, 9500 Gilman Drive, La Jolla, California 92093-0375 Correspondence: [email protected] The prosurvival transcription factor NF-kB specifically binds promoter DNA to activate target gene expression. NF-kB is regulated through interactions with IkB inhibitor proteins. Active proteolysis of these IkB proteins is, in turn, under the control of the IkB kinase complex (IKK). Together, these three molecules form the NF-kB signaling module. Studies aimed at charac- terizing the molecular mechanisms of NF-kB, IkB, and IKK in terms of their three-dimen- sional structures have lead to a greater understanding of this vital transcription factor system. F-kB is a master transcription factor that from the perspective of their three-dimensional Nresponds to diverse cell stimuli by activat- structures. ing the expression of stress response genes. Multiple signals, including cytokines, growth factors, engagement of the T-cell receptor, and NF-kB bacterial and viral products, induce NF-kB Introduction to NF-kB transcriptional activity (Hayden and Ghosh 2008). A point of convergence for the myriad NF-kB was discovered in the laboratory of of NF-kB inducing signals is the IkB kinase David Baltimore as a nuclear activity with bind- complex (IKK). Active IKK in turn controls ing specificity toward a ten-base-pair DNA transcription factor NF-kB by regulating pro- sequence 50-GGGACTTTCC-30 present within teolysis of the IkB inhibitor protein (Fig. -
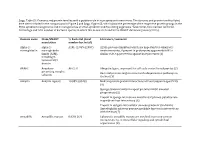
Supp. Table S2: Domains and Protein Families with a Putative Role in Host-Symbiont Interactions
Supp. Table S2: Domains and protein families with a putative role in host-symbiont interactions. The domains and protein families listed here were included in the comparisons in Figure 5 and Supp. Figure S5, which show the percentage of the respective protein groups in the Riftia symbiont metagenome and in metagenomes of other symbiotic and free-living organisms. % bacterial, total number bacterial: Percentage and total number of bacterial species in which this domain is found in the SMART database (January 2019). Domain name Pfam/SMART % bacterial (total Literature/comment annotation number bacterial) Alpha-2- alpha-2- A2M: 42.05% (2057) A2Ms: protease inhibitors which are important for eukaryotic macroglobulin macroglobulin innate immunity, if present in prokaryotes apparently fulfill a family (A2M), similar role, e.g. protection against host proteases (1) including N- terminal MG1 domain ANAPC Anaphase- APC2: 0 Ubiquitin ligase, important for cell cycle control in eukaryotes (2) promoting complex Bacterial proteins might interact with ubiquitination pathways in subunits the host (3) Ankyrin Ankyrin repeats 10.88% (8348) Mediate protein-protein interactions without sequence specificity (4) Sponge symbiont ankyrin-repeat proteins inhibit amoebal phagocytosis (5) Present in sponge microbiome metatranscriptomes, putative role in symbiont-host interactions (6) Present in obligate intracellular amoeba symbiont Candidatus Amoebophilus asiaticus genome, probable function in interactions with the host (7) Armadillo Armadillo repeats 0.83% (67) -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

Strategies and Challenges for the Next Generation of Therapeutic Antibodies
FOCUS ON THERAPEUTIC ANTIBODIES PERSPECTIVES ‘validated targets’, either because prior anti- TIMELINE bodies have clearly shown proof of activity in humans (first-generation approved anti- Strategies and challenges for the bodies on the market for clinically validated targets) or because a vast literature exists next generation of therapeutic on the importance of these targets for the disease mechanism in both in vitro and in vivo pharmacological models (experi- antibodies mental validation; although this does not necessarily equate to clinical validation). Alain Beck, Thierry Wurch, Christian Bailly and Nathalie Corvaia Basically, the strategy consists of develop- ing new generations of antibodies specific Abstract | Antibodies and related products are the fastest growing class of for the same antigens but targeting other therapeutic agents. By analysing the regulatory approvals of IgG-based epitopes and/or triggering different mecha- biotherapeutic agents in the past 10 years, we can gain insights into the successful nisms of action (second- or third-generation strategies used by pharmaceutical companies so far to bring innovative drugs to antibodies, as discussed below) or even the market. Many challenges will have to be faced in the next decade to bring specific for the same epitopes but with only one improved property (‘me better’ antibod- more efficient and affordable antibody-based drugs to the clinic. Here, we ies). This validated approach has a high discuss strategies to select the best therapeutic antigen targets, to optimize the probability of success, but there are many structure of IgG antibodies and to design related or new structures with groups working on this class of target pro- additional functions. -

2010.01) C07k 14/74 (2006.01
( (51) International Patent Classification: su 215 123 (CN). LIN, Yanni; F2, Building B20, Sangt- C07K 19/00 (2006.0 1) C12N 5/0783 (20 10.01) ian Street 218, Suzhou Industrial Park, Suzhou, Jiangsu C07K 14/74 (2006.01) A61K 35/1 7 (2015.01) 215123 (CN). ZHENG, Xiaocui; F2, Building B20, Sangt- C12N 15/62 (2006.01) A61P 35/00 (2006.01) ian Street 218, Suzhou Industrial Park, Suzhou, Jiangsu 215123 (CN). KONG, Hongmei; F2, Building B20, Sangt- (21) International Application Number: ian Street 218, Suzhou Industrial Park, Suzhou, Jiangsu PCT/ CN2020/ 1081 16 215123 (CN). WANG, Wenbo; F2, Building B20, Sangtian (22) International Filing Date: Street 218, Suzhou Industrial Park, Suzhou, Jiangsu 215 123 10 August 2020 (10.08.2020) (CN). FENG, Aihua; F2, Building B20, Sangtian Street 218, Suzhou Industrial Park, Suzhou, Jiangsu 215 123 (CN). (25) Filing Language: English (74) Agent: J. Z. M. C. PATENT AND TRADEMARK LAW (26) Publication Language: English OFFICE (GENERAL PARTNERSHIP); YU, Mingwei, (30) Priority Data: Room 5022, No. 335, Guo Ding Road, Yang Pu District, 201910746355.8 13 August 2019 (13.08.2019) CN Shanghai 200433 (CN). PCT/ CN20 19/124321 (81) Designated States (unless otherwise indicated, for every 10 December 2019 (10. 12.2019) CN kind of national protection av ailable) . AE, AG, AL, AM, (71) Applicant: CURE GENETICS CO., LTD. [CN/CN]; AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, Biobay A4-5 10, Xinghu Street 218, Suzhou Industrial Park, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, Suzhou, Jiangsu 215 123 (CN). -

EURL ECVAM Recommendation on Non-Animal-Derived Antibodies
EURL ECVAM Recommendation on Non-Animal-Derived Antibodies EUR 30185 EN Joint Research Centre This publication is a Science for Policy report by the Joint Research Centre (JRC), the European Commission’s science and knowledge service. It aims to provide evidence-based scientific support to the European policymaking process. The scientific output expressed does not imply a policy position of the European Commission. Neither the European Commission nor any person acting on behalf of the Commission is responsible for the use that might be made of this publication. For information on the methodology and quality underlying the data used in this publication for which the source is neither Eurostat nor other Commission services, users should contact the referenced source. EURL ECVAM Recommendations The aim of a EURL ECVAM Recommendation is to provide the views of the EU Reference Laboratory for alternatives to animal testing (EURL ECVAM) on the scientific validity of alternative test methods, to advise on possible applications and implications, and to suggest follow-up activities to promote alternative methods and address knowledge gaps. During the development of its Recommendation, EURL ECVAM typically mandates the EURL ECVAM Scientific Advisory Committee (ESAC) to carry out an independent scientific peer review which is communicated as an ESAC Opinion and Working Group report. In addition, EURL ECVAM consults with other Commission services, EURL ECVAM’s advisory body for Preliminary Assessment of Regulatory Relevance (PARERE), the EURL ECVAM Stakeholder Forum (ESTAF) and with partner organisations of the International Collaboration on Alternative Test Methods (ICATM). Contact information European Commission, Joint Research Centre (JRC), Chemical Safety and Alternative Methods Unit (F3) Address: via E. -

WSB1: from Homeostasis to Hypoxia Moinul Haque1,2,3, Joseph Keith Kendal1,2,3, Ryan Matthew Macisaac1,2,3 and Douglas James Demetrick1,2,3,4*
Haque et al. Journal of Biomedical Science (2016) 23:61 DOI 10.1186/s12929-016-0270-3 REVIEW Open Access WSB1: from homeostasis to hypoxia Moinul Haque1,2,3, Joseph Keith Kendal1,2,3, Ryan Matthew MacIsaac1,2,3 and Douglas James Demetrick1,2,3,4* Abstract The wsb1 gene has been identified to be important in developmental biology and cancer. A complex transcriptional regulation of wsb1 yields at least three functional transcripts. The major expressed isoform, WSB1 protein, is a substrate recognition protein within an E3 ubiquitin ligase, with the capability to bind diverse targets and mediate ubiquitinylation and proteolytic degradation. Recent data suggests a new role for WSB1 as a component of a neuroprotective pathway which results in modification and aggregation of neurotoxic proteins such as LRRK2 in Parkinson’s Disease, via an unusual mode of protein ubiquitinylation. WSB1 is also involved in thyroid hormone homeostasis, immune regulation and cellular metabolism, particularly glucose metabolism and hypoxia. In hypoxia, wsb1 is a HIF-1 target, and is a regulator of the degradation of diverse proteins associated with the cellular response to hypoxia, including HIPK2, RhoGDI2 and VHL. Major roles are to both protect HIF-1 function through degradation of VHL, and decrease apoptosis through degradation of HIPK2. These activities suggest a role for wsb1 in cancer cell proliferation and metastasis. As well, recent work has identified a role for WSB1 in glucose metabolism, and perhaps in mediating the Warburg effect in cancer cells by maintaining the function of HIF1. Furthermore, studies of cancer specimens have identified dysregulation of wsb1 associated with several types of cancer, suggesting a biologically relevant role in cancer development and/or progression. -

Monobodies and Other Synthetic Binding Proteins for Expanding Protein Science
Reviews Protein Science DOI 10.1002/pro.3148 Monobodies and Other Synthetic Binding Proteins for Expanding Protein Science Fern Sha,1,4 Gabriel Salzman,1 Ankit Gupta1,2 and Shohei Koide1,2,3* 1Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA 2Perlmutter Cancer Center, New York University Langone Medical Center, New York, NY 10016, USA 3Department of Biochemistry and Molecular Pharmacology New York University School of Medicine, New York, NY 10016, USA 4Present address, Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA 20148, USA *Correspondence to: Shohei Koide, Perlmutter Cancer Center, New York University Langone Medical Center, New York, NY 10016, USA. E-mail: [email protected] Abstract Synthetic binding proteins are constructed using non-antibody molecular scaffolds. Over the last two decades, in-depth structural and functional analyses of synthetic binding proteins have improved combinatorial library designs and selection strategies, which have resulted in potent platforms that consistently generate binding proteins to diverse targets with affinity and specificity that rival those of antibodies. Favorable attributes of synthetic binding proteins, such as small size, freedom from disulfide bond formation and ease of making fusion proteins, have enabled their unique applications in protein science, cell biology and beyond. Here, we review recent studies that illustrate how synthetic binding proteins are powerful probes that can directly link structure and function, -

Highly Potent Anti-SARS-Cov-2 Multi-Darpin Therapeutic Candidates Marcel Walser1,*, Sylvia Rothenberger2,3,*, Daniel L
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.25.256339; this version posted August 26, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Highly potent anti-SARS-CoV-2 multi-DARPin therapeutic candidates Marcel Walser1,*, Sylvia Rothenberger2,3,*, Daniel L. Hurdiss4,5,*, Anja Schlegel1, Valérie Calabro1, Simon Fontaine1, Denis Villemagne1, Maria Paladino1, Tanja Hospodarsch1, Alexandra Neculcea1, Andreas Cornelius1, Patricia Schildknecht1, Mirela Matzner1, Martin Hänggi1, Marco Franchini1, Yvonne 5 Kaufmann1, Iris Schlegel1,, Chloe Iss1, Thamar Loser1, Susanne Mangold1, Christel Herzog1, Dieter Schiegg1, Christian Reichen1, Filip Radom1, Andreas Bosshart1, Andreas Lehmann1, Micha A. Haeuptle1, Alexander Zürcher1, Toni Vagt1, Gabriel Sigrist1, Marcel Straumann 1, Karl Proba1, Niina Veitonmäki1, Keith M. Dawson1, Christof Zitt1, Jennifer Mayor2,3, Sarah Ryter2, Heyrhyoung Lyoo4, Chunyan Wang4, Wentao Li4, Ieva Drulyte6, H. Kaspar Binz7, Leon de Waal8, Koert J. Stittelaar8, Seth Lewis1, Daniel 10 Steiner1, Frank J.M. van Kuppeveld4, Olivier Engler2, Berend-Jan Bosch4, Michael T. Stumpp1,9, Patrick Amstutz 1 *These authors contributed equally to this work 1Molecular Partners AG, Wagistrasse 14, 8952 Zurich-Schlieren, Switzerland 15 2Spiez Laboratory, Austrasse, 3700 Spiez, Switzerland 3Institute of Microbiology, University Hospital Center -

(12) United States Patent (10) Patent No.: US 9,499,622 B2 Cheong Et Al
USOO9499622B2 (12) United States Patent (10) Patent No.: US 9,499,622 B2 Cheong et al. (45) Date of Patent: Nov. 22, 2016 (54) ANTI-EGFR/ANTI-HER2 BISPECIFIC 5,821,337 A 10, 1998 Carter et al. ANTIBODIES WITH ANT-EGFR DARPNS 6,165,464 A 12/2000 Hudziak et al. 7,417,130 B2 8/2008 Stumpp et al. 8,071,730 B2 12/2011 Goetsch et al. (71) Applicant: Samsung Electronics Co., Ltd., 8, 110,653 B2 2/2012 Stumpp et al. Suwon-si, Gyeonggi-do (KR) 8,329,873 B2 12/2012 Adams et al. 8,524,244 B2 9/2013 Camphausen et al. (72) Inventors: Kwang Ho Cheong, Seoul (KR): 9,234,028 B2 1/2016 Camphausen et al. Seung Hyun Lee, Suwon-si (KR): 9,359.440 B2 * 6/2016 Cheong ................ CO7K 14,705 Powei Lin, Hwaseong-si (KR); Saet 2009/0010840 A1 1/2009 Adams et al. Byoul Lee, Seoul (KR); Jae Woong 2010/0178298 A1 7/2010 Lindhofer 2012fOO2O966 A1* 1/2012 Barbas, III ........... CO7K 14,515 Hwang, Seoul (KR) 424,134.1 2012/0156191 A1 6/2012 Goetsch et al. (73) Assignee: SAMSUNGELECTRONICS CO., 2012,0277143 A1 1 1/2012 Jacobs et al. LTD., Suwon-Si (KR) 2013, OO17200 A1 1/2013 Scheer et al. 2015.OO30596 A1* 1/2015 Cheong ................ CO7K 14,705 (*) Notice: Subject to any disclaimer, the term of this 424,134.1 patent is extended or adjusted under 35 U.S.C. 154(b) by 0 days. FOREIGN PATENT DOCUMENTS EP 1332.209 B2 11/2009 (21) Appl.