Structure and Function of Primase Repb Encoded by Broad-Host
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Specific Binding of Transposase to Terminal Inverted Repeats Of
Proc. Natl. Acad. Sci. USA Vol. 84, pp. 8220-8224, December 1987 Biochemistry Specific binding of transposase to terminal inverted repeats of transposable element Tn3 (transposon/DNA binding protein/DNase I "footprinting") HITOSHI ICHIKAWA*, KAORU IKEDA*, WILLIAM L. WISHARTt, AND EIICHI OHTSUBO*t *Institute of Applied Microbiology, University of Tokyo, Bunkyo-ku, Yayoi 1-1-1, Tokyo 113, Japan; and tDepartment of Molecular Biology, Princeton University, Princeton, NJ 08544 Communicated by Norman Davidson, July 27, 1987 ABSTRACT Tn3 transposase, which is required for trans- containing the Tn3 terminal regions when 8 mM ATP is position of Tn3, has been purified by a low-ionic-strength- present (14). precipitation method. Using a nitrocellulose filter binding In this paper, we study the interaction of Tn3 transposase, assay, we have shown that transposase binds to any restriction which has been purified by a low-ionic-strength-precipitation fragment. However, binding of the transposase to specific method, with DNA. We demonstrate that the purified fragments containing the terminal inverted repeat sequences of transposase is an ATP-independent DNA binding protein, Tn3 can be demonstrated by treatment of transposase-DNA which has interesting properties that may be involved in the complexes with heparin, which effectively removes the actual transposition event of Tn3. transposase bound to the other nonspecific fragments at pH 5-6. DNase I "footprinting" analysis showed that the MATERIALS AND METHODS transposase protects an inner 25-base-pair region of the 38- base-pair terminal inverted repeat sequence of Tn3. This Bacterial Strains and Plasmids. The bacterial strains used protection is not dependent on pH. -

A Sleeping Beauty Mutagenesis Screen Reveals a Tumor Suppressor Role for Ncoa2/Src-2 in Liver Cancer
A Sleeping Beauty mutagenesis screen reveals a tumor suppressor role for Ncoa2/Src-2 in liver cancer Kathryn A. O’Donnella,b,1,2, Vincent W. Kengc,d,e, Brian Yorkf, Erin L. Reinekef, Daekwan Seog, Danhua Fanc,h, Kevin A. T. Silversteinc,h, Christina T. Schruma,b, Wei Rose Xiea,b,3, Loris Mularonii,j, Sarah J. Wheelani,j, Michael S. Torbensonk, Bert W. O’Malleyf, David A. Largaespadac,d,e, and Jef D. Boekea,b,i,2 Departments of aMolecular Biology and Genetics, iOncology, jDivision of Biostatistics and Bioinformatics, and kPathology, and bThe High Throughput Biology Center, The Johns Hopkins University School of Medicine, Baltimore, MD 21205; cMasonic Cancer Center, dDepartment of Genetics, Cell Biology, and Development, eCenter for Genome Engineering, and hBiostatistics and Bioinformatics Core, University of Minnesota, Minneapolis, MN 55455; fDepartment of Molecular and Cellular Biology, Baylor College of Medicine, Houston, TX 77030; and gLaboratory of Experimental Carcinogenesis, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892 AUTHOR SUMMARY Emerging data from cancer ge- functions are altered. This ap- fi nome-sequencing studies have Sleeping Beauty (SB) transposase proach identi ed at least 16 demonstrated that human genes/loci that contribute to liver tumors exhibit tremendous Transposon array with gene trap (GT) tumor development. complexity and heterogeneity in We next validated that the the number and nature of iden- genes identified in the SB screen tified mutations (1). Based on contribute to tumor initiation these findings, there is an in- SB mobilization and/or progression using in vitro creasing need for in vivo vali- and in vivo cancer model sys- dation of genes whose altered tems. -

RNA As a Source of Transposase for Sleeping Beauty-Mediated Gene Insertion and Expression in Somatic Cells and Tissues
doi:10.1016/j.ymthe.2005.10.014 SHORT COMMUNICATION RNA as a Source of Transposase for Sleeping Beauty-Mediated Gene Insertion and Expression in Somatic Cells and Tissues Andrew Wilber,1 Joel L. Frandsen,1 Jennifer L. Geurts,1 David A. Largaespada,1 Perry B. Hackett,1,2 and R. Scott McIvor1,* 1The Arnold and Mabel Beckman Center for Transposon Research, Gene Therapy Program, Institute of Human Genetics, and Department of Genetics, Cell Biology and Development, University of Minnesota, Minneapolis, MN 55455, USA 2Discovery Genomics, Inc., Minneapolis, MN 55455, USA *To whom correspondence and reprint requests should be addressed at The Department of Genetics, Cell Biology, and Development, 6-160 Jackson Hall, 321 Church Street SE, University of Minnesota, Minneapolis, MN 55455, USA. Fax: +1 612 625 9810. E-mail: [email protected]. Available online 20 December 2005 Sleeping Beauty (SB) is a DNA transposon capable of mediating gene insertion and long-term expression in vertebrate cells when co-delivered with a source of transposase. In all previous reports of SB-mediated gene insertion in somatic cells, the transposase component has been provided by expression of a co-delivered DNA molecule that has the potential for integration into the host cell genome. Integration and continued expression of a gene encoding SB transposase could be problematic if it led to transposon re-mobilization and re-integration. We addressed this potential problem by supplying the transposase-encoding molecule in the form of mRNA. We show that transposase-encoding mRNA can effectively mediate transposition in vitro in HT1080 cells and in vivo in mouse liver following co-delivery with a recoverable transposon or with a luciferase transposon. -

Licensing RNA-Guided Genome Defense
TIBS-1023; No. of Pages 10 Review Recognizing the enemy within: licensing RNA-guided genome defense Phillip A. Dumesic and Hiten D. Madhani Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94158, USA How do cells distinguish normal genes from transpo- RNAi-related RNA silencing pathways are deeply con- sons? Although much has been learned about RNAi- served genome defense mechanisms that act in organisms related RNA silencing pathways responsible for genome from protist to human to recognize and suppress transpo- defense, this fundamental question remains. The litera- sons [8]. In all of these pathways, 20–30 nucleotide small ture points to several classes of mechanisms. In some RNAs act in complex with Argonaute family proteins to cases, double-stranded RNA (dsRNA) structures pro- silence complementary transcripts. Depending on the duced by transposon inverted repeats or antisense inte- pathway and context, silencing proceeds by a variety of gration trigger endogenous small interfering RNA mechanisms, which include RNA degradation, translation- (siRNA) biogenesis. In other instances, DNA features al repression, and the establishment of repressive histone associated with transposons – such as their unusual modifications [9,10]. The pathways also differ in the means copy number, chromosomal arrangement, and/or chro- by which they generate small RNAs. In the canonical RNAi matin environment – license RNA silencing. Finally, re- pathway, RNase-III-type Dicer enzymes convert long cent studies have identified improper transcript dsRNA into small interfering RNA (siRNA). In contrast, processing events, such as stalled pre-mRNA splicing, PIWI-interacting RNA (piRNA) pathways do not require as signals for siRNA production. -

Exploring the Non-Canonical Functions of Metabolic Enzymes Peiwei Huangyang1,2 and M
© 2018. Published by The Company of Biologists Ltd | Disease Models & Mechanisms (2018) 11, dmm033365. doi:10.1242/dmm.033365 REVIEW SPECIAL COLLECTION: CANCER METABOLISM Hidden features: exploring the non-canonical functions of metabolic enzymes Peiwei Huangyang1,2 and M. Celeste Simon1,3,* ABSTRACT A key finding from studies of metabolic enzymes is the existence The study of cellular metabolism has been rigorously revisited over the of mechanistic links between their nuclear localization and the past decade, especially in the field of cancer research, revealing new regulation of transcription. By modulating gene expression, insights that expand our understanding of malignancy. Among these metabolic enzymes themselves facilitate adaptation to rapidly insights isthe discovery that various metabolic enzymes have surprising changing environments. Furthermore, they can directly shape a ’ activities outside of their established metabolic roles, including in cell s epigenetic landscape (Kaelin and McKnight, 2013). the regulation of gene expression, DNA damage repair, cell cycle Strikingly, several metabolic enzymes exert completely distinct progression and apoptosis. Many of these newly identified functions are functions in different cellular compartments. Nuclear fructose activated in response to growth factor signaling, nutrient and oxygen bisphosphate aldolase, for example, directly interacts with RNA ́ availability, and external stress. As such, multifaceted enzymes directly polymerase III to control transcription (Ciesla et al., 2014), -

Catalytic Domain of Plasmid Pad1 Relaxase Trax Defines a Group Of
Catalytic domain of plasmid pAD1 relaxase TraX defines a group of relaxases related to restriction endonucleases María Victoria Franciaa,1, Don B. Clewellb,c, Fernando de la Cruzd, and Gabriel Moncaliánd aServicio de Microbiología, Hospital Universitario Marqués de Valdecilla e Instituto de Formación e Investigación Marqués de Valdecilla, Santander 39008, Spain; bDepartment of Biologic and Materials Sciences, University of Michigan School of Dentistry, Ann Arbor, MI 48109; cDepartment of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, MI 48109; and dDepartamento de Biología Molecular e Instituto de Biomedicina y Biotecnología de Cantabria, Universidad de Cantabria–Consejo Superior de Investigaciones Científicas–Sociedad para el Desarrollo Regional de Cantabria, Santander 39011, Spain Edited by Roy Curtiss III, Arizona State University, Tempe, AZ, and approved July 9, 2013 (received for review May 30, 2013) Plasmid pAD1 is a 60-kb conjugative element commonly found in known relaxases show two characteristic sequence motifs, motif I clinical isolates of Enterococcus faecalis. The relaxase TraX and the containing the catalytic Tyr residue (which covalently attaches to primary origin of transfer oriT2 are located close to each other and the 5′ end of the cleaved DNA) and motif III with the His-triad have been shown to be essential for conjugation. The oriT2 site essential for relaxase activity (facilitating the cleavage reaction contains a large inverted repeat (where the nic site is located) by activation of the catalytic Tyr). This His-triad has been used as adjacent to a series of short direct repeats. TraX does not show a relaxase diagnostic signature (12). fi any of the typical relaxase sequence motifs but is the prototype of Plasmid pAD1 relaxase, TraX, was identi ed (9) and shown to oriT2 a unique family of relaxases (MOB ). -

Arthur Kornberg Discovered (The First) DNA Polymerase Four
Arthur Kornberg discovered (the first) DNA polymerase Using an “in vitro” system for DNA polymerase activity: 1. Grow E. coli 2. Break open cells 3. Prepare soluble extract 4. Fractionate extract to resolve different proteins from each other; repeat; repeat 5. Search for DNA polymerase activity using an biochemical assay: incorporate radioactive building blocks into DNA chains Four requirements of DNA-templated (DNA-dependent) DNA polymerases • single-stranded template • deoxyribonucleotides with 5’ triphosphate (dNTPs) • magnesium ions • annealed primer with 3’ OH Synthesis ONLY occurs in the 5’-3’ direction Fig 4-1 E. coli DNA polymerase I 5’-3’ polymerase activity Primer has a 3’-OH Incoming dNTP has a 5’ triphosphate Pyrophosphate (PP) is lost when dNMP adds to the chain E. coli DNA polymerase I: 3 separable enzyme activities in 3 protein domains 5’-3’ polymerase + 3’-5’ exonuclease = Klenow fragment N C 5’-3’ exonuclease Fig 4-3 E. coli DNA polymerase I 3’-5’ exonuclease Opposite polarity compared to polymerase: polymerase activity must stop to allow 3’-5’ exonuclease activity No dNTP can be re-made in reversed 3’-5’ direction: dNMP released by hydrolysis of phosphodiester backboneFig 4-4 Proof-reading (editing) of misincorporated 3’ dNMP by the 3’-5’ exonuclease Fidelity is accuracy of template-cognate dNTP selection. It depends on the polymerase active site structure and the balance of competing polymerase and exonuclease activities. A mismatch disfavors extension and favors the exonuclease.Fig 4-5 Superimposed structure of the Klenow fragment of DNA pol I with two different DNAs “Fingers” “Thumb” “Palm” red/orange helix: 3’ in red is elongating blue/cyan helix: 3’ in blue is getting edited Fig 4-6 E. -

Teacher Notes Science Nspired
DNA Replication TEACHER NOTES SCIENCE NSPIRED Science Objectives In this lesson, students will • Explore how the structure of DNA supports its semi-conservative replication • Identify the name and function of several key enzymes in DNA replication • Recognize the function of Okazaki fragments in DNA replication Vocabulary TI-Nspire™ Technology Skills: • Double helix • Leading strand • Download a TI-Nspire • Lagging strand • Polymerase I • Polymerase III • Helicase document • Base pairs • Okazaki Fragments • Open a document • Primase • Move between pages • Open Directions Box About the Lesson • Explore ‘Hot Spots’ • Using three simulations, students will interact with DNA replication to explore semi-conservative replication and identify Tech Tips: specific enzymes and their roles in replication. Assessments are Make sure that students know embedded in the activity to engage discussion and gauge how to move between pages by learning. pressing /¡ (left arrow) and • As a result, students will: /¢ (right arrow). • Learn the basic functions of the following DNA replication enzymes: helicase, primase, ligase, polymerase I and III. Lesson Materials: • Learn how the double helix reproduces in a semi- Student Activity • DNA_Replication_Student.do conservative fashion c • Learn the role and function of Okazaki fragments in • DNA_Replication_Student.pd replication of the lagging strand. f TI-Nspire document TI-Nspire™ Navigator™ • DNA_Replication.tns Not required. If using Navigator: • Send out the DNA_Replication.tns file. • Monitor student progress -
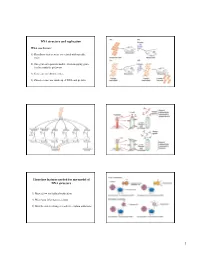
DNA Structure and Replication Three Key Features Needed for Any Model
DNA structure and replication What was known? 1) Hereditary factors were associated with specific traits 2) One-gene-one-protein model - from mapping genes for biosynthetic pathways 3) Genes are on chromosomes 4) Chromosomes are made up of DNA and protein Three key features needed for any model of DNA structure 1) Must allow for faithful replication 2) Must have information content 3) Must be able to change in order to explain mutations 1 Rosalind Franklin James Watson Francis Crick X-ray diffraction Maurice Wilkins Photograph of B-DNA Linus Pauling Nature 171, 737-738 (1953) © Macmillan Publishers Ltd. Molecular structure of Nucleic Acids WATSON, J. D. & CRICK, F. H. C. Features of the double helix A Structure for Deoxyribose Nucleic Acid 1) Two parallel strands We wish to suggest a structure for the salt of deoxyribose nucleic acid (D.N.A.). This structure has novel features which are of considerable biological interest………… 2) Bases held together by H-bonds 3) Phosphodiester backbone 4) The base is attached to the position 1 on the sugar 5) Base pair stack - provides Figure 1. This figure is purely diagrammatic. The two ribbons symbolize the two phophate-sugar chains, and the horizonal rods stability the pairs of bases holding the chains together. The vertical line marks the fibre axis. 6) Contains a major and …………….It has not escaped our notice that the specific pairing we minor groove have postulated immediately suggests a possible copying mechanism for the genetic material. Three key features needed for any model of DNA structure -

Processivity of DNA Polymerases: Two Mechanisms, One Goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2
Minireview 121 Processivity of DNA polymerases: two mechanisms, one goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2 Replicative DNA polymerases are highly processive Processive DNA synthesis by cellular replicases and the enzymes that polymerize thousands of nucleotides without bacteriophage T4 replicase dissociating from the DNA template. The recently Until recently, the only mechanism for high processivity determined structure of the Escherichia coli bacteriophage that was understood in detail was that utilized by cellular T7 DNA polymerase suggests a unique mechanism that replicases and the replicase of bacteriophage T4. This underlies processivity, and this mechanism may generalize mechanism involves a ring-shaped protein called a ‘DNA to other replicative polymerases. sliding clamp’ that encircles the DNA and tethers the polymerase catalytic unit to the DNA [3,4]. The three- Addresses: 1Department of Molecular Biology, Memorial Sloan- dimensional structures of several sliding clamps have been Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, 2 determined: the eukaryotic proliferating cell nuclear USA and Laboratory of DNA Replication, Howard Hughes Medical β Institute, The Rockefeller University, 1230 York Avenue, New York, NY antigen (PCNA) [5,6]; the subunit of the prokaryotic 10021, USA. DNA polymerase III [7]; and the bacteriophage T4 gene 45 protein (gp45) (J Kuriyan, personal communication) *Corresponding author. (Figure 1). The overall structure of these clamps is very E-mail: [email protected] similar; the PCNA, β subunit and gp45 rings are super- Structure 15 February 1998, 6:121–125 imposable [8]. Each ring has similar dimensions and a http://biomednet.com/elecref/0969212600600121 central cavity large enough to accommodate duplex DNA (Figure 1). -

The Obscure World of Integrative and Mobilizable Elements Gérard Guédon, Virginie Libante, Charles Coluzzi, Sophie Payot-Lacroix, Nathalie Leblond-Bourget
The obscure world of integrative and mobilizable elements Gérard Guédon, Virginie Libante, Charles Coluzzi, Sophie Payot-Lacroix, Nathalie Leblond-Bourget To cite this version: Gérard Guédon, Virginie Libante, Charles Coluzzi, Sophie Payot-Lacroix, Nathalie Leblond-Bourget. The obscure world of integrative and mobilizable elements: Highly widespread elements that pirate bacterial conjugative systems. Genes, MDPI, 2017, 8 (11), pp.337. 10.3390/genes8110337. hal- 01686871 HAL Id: hal-01686871 https://hal.archives-ouvertes.fr/hal-01686871 Submitted on 26 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License G C A T T A C G G C A T genes Review The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems Gérard Guédon *, Virginie Libante, Charles Coluzzi, Sophie Payot and Nathalie Leblond-Bourget * ID DynAMic, Université de Lorraine, INRA, 54506 Vandœuvre-lès-Nancy, France; [email protected] (V.L.); [email protected] (C.C.); [email protected] (S.P.) * Correspondence: [email protected] (G.G.); [email protected] (N.L.-B.); Tel.: +33-037-274-5142 (G.G.); +33-037-274-5146 (N.L.-B.) Received: 12 October 2017; Accepted: 15 November 2017; Published: 22 November 2017 Abstract: Conjugation is a key mechanism of bacterial evolution that involves mobile genetic elements. -

Primer Release Is the Rate-Limiting Event in Lagging-Strand Synthesis
Primer release is the rate-limiting event in lagging- strand synthesis mediated by the T7 replisome Alfredo J. Hernandeza, Seung-Joo Leea, and Charles C. Richardsona,1 aDepartment of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115 Contributed by Charles C. Richardson, April 18, 2016 (sent for review December 30, 2015; reviewed by Nicholas E. Dixon and I. Robert Lehman) DNA replication occurs semidiscontinuously due to the antiparallel but also for enabling the use of short oligoribonucleotides by T7 DNA strands and polarity of enzymatic DNA synthesis. Although DNA polymerase. Critically, the primase domain also fulfills two the leading strand is synthesized continuously, the lagging strand additional roles apart from primer synthesis: it prevents disso- is synthesized in small segments designated Okazaki fragments. ciation of the extremely short tetramer, stabilizing it with the Lagging-strand synthesis is a complex event requiring repeated template, and it secures it in the polymerase active site (10, 12). cycles of RNA primer synthesis, transfer to the lagging-strand Here we show that the rate-limiting step in initiation of Okazaki polymerase, and extension effected by cooperation between DNA fragments by the T7 replisome is primer release from the primase primase and the lagging-strand polymerase. We examined events domain of gp4. In the absence of gp2.5, an additional step, distinct controlling Okazaki fragment initiation using the bacteriophage from primer release, also limits primer extension. The presence of T7 replication system. Primer utilization by T7 DNA polymerase is gp2.5 promotes efficient primer formation and primer utilization. slower than primer formation.