Bioinformatic Characterization of Angiotensin-Converting Enzyme 2, the Entry Receptor for SARS-Cov-2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ACE2 Interaction Networks in COVID-19: a Physiological Framework for Prediction of Outcome in Patients with Cardiovascular Risk Factors
Journal of Clinical Medicine Article ACE2 Interaction Networks in COVID-19: A Physiological Framework for Prediction of Outcome in Patients with Cardiovascular Risk Factors Zofia Wicik 1,2 , Ceren Eyileten 2, Daniel Jakubik 2,Sérgio N. Simões 3, David C. Martins Jr. 1, Rodrigo Pavão 1, Jolanta M. Siller-Matula 2,4,* and Marek Postula 2 1 Centro de Matemática, Computação e Cognição, Universidade Federal do ABC, Santo Andre 09606-045, Brazil; zofi[email protected] (Z.W.); [email protected] (D.C.M.J.); [email protected] (R.P.) 2 Department of Experimental and Clinical Pharmacology, Medical University of Warsaw, Center for Preclinical Research and Technology CEPT, 02-091 Warsaw, Poland; [email protected] (C.E.); [email protected] (D.J.); [email protected] (M.P.) 3 Federal Institute of Education, Science and Technology of Espírito Santo, Serra, Espírito Santo 29056-264, Brazil; [email protected] 4 Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, 1090 Vienna, Austria * Correspondence: [email protected]; Tel.: +43-1-40400-46140; Fax: +43-1-40400-42160 Received: 9 October 2020; Accepted: 17 November 2020; Published: 21 November 2020 Abstract: Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (coronavirus disease 2019; COVID-19) is associated with adverse outcomes in patients with cardiovascular disease (CVD). The aim of the study was to characterize the interaction between SARS-CoV-2 and Angiotensin-Converting Enzyme 2 (ACE2) functional networks with a focus on CVD. Methods: Using the network medicine approach and publicly available datasets, we investigated ACE2 tissue expression and described ACE2 interaction networks that could be affected by SARS-CoV-2 infection in the heart, lungs and nervous system. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

Pancancer Progression Human Vjune2017
Gene Symbol Accession Alias/Prev Symbol Official Full Name AAMP NM_001087.3 - angio-associated, migratory cell protein ABI3BP NM_015429.3 NESHBP|TARSH ABI family, member 3 (NESH) binding protein ACHE NM_000665.3 ACEE|ARACHE|N-ACHE|YT acetylcholinesterase ACTG2 NM_001615.3 ACT|ACTA3|ACTE|ACTL3|ACTSG actin, gamma 2, smooth muscle, enteric ACVR1 NM_001105.2 ACTRI|ACVR1A|ACVRLK2|ALK2|FOP|SKR1|TSRI activin A receptor, type I ACVR1C NM_145259.2 ACVRLK7|ALK7 activin A receptor, type IC ACVRL1 NM_000020.1 ACVRLK1|ALK-1|ALK1|HHT|HHT2|ORW2|SKR3|TSR-I activin A receptor type II-like 1 ADAM15 NM_207195.1 MDC15 ADAM metallopeptidase domain 15 ADAM17 NM_003183.4 ADAM18|CD156B|CSVP|NISBD|TACE ADAM metallopeptidase domain 17 ADAM28 NM_014265.4 ADAM 28|ADAM23|MDC-L|MDC-Lm|MDC-Ls|MDCL|eMDC II|eMDCII ADAM metallopeptidase domain 28 ADAM8 NM_001109.4 CD156|MS2 ADAM metallopeptidase domain 8 ADAM9 NM_001005845.1 CORD9|MCMP|MDC9|Mltng ADAM metallopeptidase domain 9 ADAMTS1 NM_006988.3 C3-C5|METH1 ADAM metallopeptidase with thrombospondin type 1 motif, 1 ADAMTS12 NM_030955.2 PRO4389 ADAM metallopeptidase with thrombospondin type 1 motif, 12 ADAMTS8 NM_007037.4 ADAM-TS8|METH2 ADAM metallopeptidase with thrombospondin type 1 motif, 8 ADAP1 NM_006869.2 CENTA1|GCS1L|p42IP4 ArfGAP with dual PH domains 1 ADD1 NM_001119.4 ADDA adducin 1 (alpha) ADM2 NM_001253845.1 AM2|dJ579N16.4 adrenomedullin 2 ADRA2B NM_000682.4 ADRA2L1|ADRA2RL1|ADRARL1|ALPHA2BAR|alpha-2BAR adrenoceptor alpha 2B AEBP1 NM_001129.3 ACLP AE binding protein 1 AGGF1 NM_018046.3 GPATC7|GPATCH7|HSU84971|HUS84971|VG5Q -

Appendix Table A.2.3.1 Full Table of All Chicken Proteins and Human Orthologs Pool Accession Human Human Protein Human Product Cell Angios Log2( Endo Gene Comp
Appendix table A.2.3.1 Full table of all chicken proteins and human orthologs Pool Accession Human Human Protein Human Product Cell AngioS log2( Endo Gene comp. core FC) Specific CIKL F1NWM6 KDR NP_002244 kinase insert domain receptor (a type III receptor tyrosine M 94 4 kinase) CWT Q8AYD0 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CWT Q8AYD0 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1P1Y9 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1P1Y9 CDH5 NP_001786 cadherin 5, type 2 (vascular endothelium) M 90 8.45 specific CIKL F1N871 FLT4 NP_891555 fms-related tyrosine kinase 4 M 86 -1.71 CWT O73739 EDNRA NP_001948 endothelin receptor type A M 81 -8 CIKL O73739 EDNRA NP_001948 endothelin receptor type A M 81 -8 CWT Q4ADW2 PROCR NP_006395 protein C receptor, endothelial M 80 -0.36 CIKL Q4ADW2 PROCR NP_006395 protein C receptor, endothelial M 80 -0.36 CIKL F1NFQ9 TEK NP_000450 TEK tyrosine kinase, endothelial M 77 7.3 specific CWT Q9DGN6 ECE1 NP_001106819 endothelin converting enzyme 1 M 74 -0.31 CIKL Q9DGN6 ECE1 NP_001106819 endothelin converting enzyme 1 M 74 -0.31 CWT F1NIF0 CA9 NP_001207 carbonic anhydrase IX I 74 CIKL F1NIF0 CA9 NP_001207 carbonic anhydrase IX I 74 CWT E1BZU7 AOC3 NP_003725 amine oxidase, copper containing 3 (vascular adhesion protein M 70 1) CIKL E1BZU7 AOC3 NP_003725 amine oxidase, copper containing 3 (vascular adhesion protein M 70 1) CWT O93419 COL18A1 NP_569712 collagen, type XVIII, alpha 1 E 70 -2.13 CIKL O93419 -

Saccharomyces Boulardii Enhances N-Terminal Peptide Hydrolysis in Suckling Rat Small Intestine by Endoluminal Release of a Zinc-Binding Metalloprotease
0031-3998/02/5104-0528 PEDIATRIC RESEARCH Vol. 51, No. 4, 2002 Copyright © 2002 International Pediatric Research Foundation, Inc. Printed in U.S.A. Saccharomyces boulardii Enhances N-Terminal Peptide Hydrolysis in Suckling Rat Small Intestine by Endoluminal Release of a Zinc-Binding Metalloprotease JEAN-PAUL BUTS, NADINE DE KEYSER, CATHERINE STILMANT, ETIENNE SOKAL, AND SOHEILA MARANDI Laboratory of Pediatric Gastroenterology and Nutrition, Unit of Pediatric Research, Faculty of Medicine, Université Catholique de Louvain, B-1200 Brussels, Belgium ABSTRACT Saccharomyces boulardii (S. boulardii), a biotherapeutic ited in the presence of 1 mM Zn2ϩ. Oral treatment of sucklings agent effective in acute and chronic enterocolopathies, produces with S. boulardii significantly enhanced jejunal and ileal mucosal trophic intestinal effects at least in part mediated by the endolu- leucine-aminopeptidase activities by 24 and 34%, respectively, minal release of polyamines. However, the effects of the yeast on over controls. In concordance, aminopeptidase activity was en- peptide hydrolysis have not yet been studied. The objectives of hanced in jejunal and ileal endoluminal fluid samples by 47 and this study were to assess in suckling rats the endoluminal and 105%, respectively. By use of an IgG-purified antibody raised mucosal aminopeptidase activities in response to S. boulardii against the zinc-binding domain common to metalloproteases, treatment and to analyze their related mechanisms. Peptidase the yeast aminopeptidase was immunoprecipitated and detected activities were assayed on yeast cells by using several L-amino as an heteromeric enzyme of 108 and 87-kD subunits. S. bou- acid-p-nitroanilide substrates in the pH range of 2 to 10. -

Chemical Agent and Antibodies B-Raf Inhibitor RAF265
Supplemental Materials and Methods: Chemical agent and antibodies B-Raf inhibitor RAF265 [5-(2-(5-(trifluromethyl)-1H-imidazol-2-yl)pyridin-4-yloxy)-N-(4-trifluoromethyl)phenyl-1-methyl-1H-benzp{D, }imidazol-2- amine] was kindly provided by Novartis Pharma AG and dissolved in solvent ethanol:propylene glycol:2.5% tween-80 (percentage 6:23:71) for oral delivery to mice by gavage. Antibodies to phospho-ERK1/2 Thr202/Tyr204(4370), phosphoMEK1/2(2338 and 9121)), phospho-cyclin D1(3300), cyclin D1 (2978), PLK1 (4513) BIM (2933), BAX (2772), BCL2 (2876) were from Cell Signaling Technology. Additional antibodies for phospho-ERK1,2 detection for western blot were from Promega (V803A), and Santa Cruz (E-Y, SC7383). Total ERK antibody for western blot analysis was K-23 from Santa Cruz (SC-94). Ki67 antibody (ab833) was from ABCAM, Mcl1 antibody (559027) was from BD Biosciences, Factor VIII antibody was from Dako (A082), CD31 antibody was from Dianova, (DIA310), and Cot antibody was from Santa Cruz Biotechnology (sc-373677). For the cyclin D1 second antibody staining was with an Alexa Fluor 568 donkey anti-rabbit IgG (Invitrogen, A10042) (1:200 dilution). The pMEK1 fluorescence was developed using the Alexa Fluor 488 chicken anti-rabbit IgG second antibody (1:200 dilution).TUNEL staining kits were from Promega (G2350). Mouse Implant Studies: Biopsy tissues were delivered to research laboratory in ice-cold Dulbecco's Modified Eagle Medium (DMEM) buffer solution. As the tissue mass available from each biopsy was limited, we first passaged the biopsy tissue in Balb/c nu/Foxn1 athymic nude mice (6-8 weeks of age and weighing 22-25g, purchased from Harlan Sprague Dawley, USA) to increase the volume of tumor for further implantation. -

C07k - 2021.08
CPC - C07K - 2021.08 C07K PEPTIDES (peptides in foodstuffs A23; obtaining protein compositions for foodstuffs, working-up proteins for foodstuffs A23J; preparations for medicinal purposes A61K; peptides containing beta-lactam rings C07D; cyclic dipeptides not having in their molecule any other peptide link than those which form their ring, e.g. piperazine-2,5-diones, C07D; ergot alkaloids of the cyclic peptide type C07D 519/02; macromolecular compounds having statistically distributed amino acid units in their molecules, i.e. when the preparation does not provide for a specific; but for a random sequence of the amino acid units, homopolyamides and block copolyamides derived from amino acids C08G 69/00; macromolecular products derived from proteins C08H 1/00; preparation of glue or gelatine C09H; single cell proteins, enzymes C12N; genetic engineering processes for obtaining peptides C12N 15/00; compositions for measuring or testing processes involving enzymes C12Q; investigation or analysis of biological material G01N 33/00) Relationships with other classification places An amino acid per se is classified in C07D while peptides (starting from dipeptides) are classified in C07K. Subclass C07K is a function oriented entry for the compounds themselves and does not cover the application or use of the compounds under the subclass definition. For classifying such information other entries exist, for example: preservation of bodies of humans or animals or plants or parts thereof; Biocides, e.g. as disinfectants, as pesticides, as herbicides; pest repellants or attractants; plant growth regulators are classified in A01N. Preparations for medical, dental, or toilet purposes are classified in A61K. Amino acids or derivatives thereof are classified in C07C or C07D. -

Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases
Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases Alan J. Barrett Neil D. Rawlings J. Fred Woessner Editor biographies xxi Contributors xxiii Preface xxxi Introduction ' Abbreviations xxxvii ASPARTIC PEPTIDASES Introduction 1 Aspartic peptidases and their clans 3 2 Catalytic pathway of aspartic peptidases 12 Clan AA Family Al 3 Pepsin A 19 4 Pepsin B 28 5 Chymosin 29 6 Cathepsin E 33 7 Gastricsin 38 8 Cathepsin D 43 9 Napsin A 52 10 Renin 54 11 Mouse submandibular renin 62 12 Memapsin 1 64 13 Memapsin 2 66 14 Plasmepsins 70 15 Plasmepsin II 73 16 Tick heme-binding aspartic proteinase 76 17 Phytepsin 77 18 Nepenthesin 85 19 Saccharopepsin 87 20 Neurosporapepsin 90 21 Acrocylindropepsin 9 1 22 Aspergillopepsin I 92 23 Penicillopepsin 99 24 Endothiapepsin 104 25 Rhizopuspepsin 108 26 Mucorpepsin 11 1 27 Polyporopepsin 113 28 Candidapepsin 115 29 Candiparapsin 120 30 Canditropsin 123 31 Syncephapepsin 125 32 Barrierpepsin 126 33 Yapsin 1 128 34 Yapsin 2 132 35 Yapsin A 133 36 Pregnancy-associated glycoproteins 135 37 Pepsin F 137 38 Rhodotorulapepsin 139 39 Cladosporopepsin 140 40 Pycnoporopepsin 141 Family A2 and others 41 Human immunodeficiency virus 1 retropepsin 144 42 Human immunodeficiency virus 2 retropepsin 154 43 Simian immunodeficiency virus retropepsin 158 44 Equine infectious anemia virus retropepsin 160 45 Rous sarcoma virus retropepsin and avian myeloblastosis virus retropepsin 163 46 Human T-cell leukemia virus type I (HTLV-I) retropepsin 166 47 Bovine leukemia virus retropepsin 169 48 -

Myeloid Innate Immunity Mouse Vapril2018
Official Symbol Accession Alias / Previous Symbol Official Full Name 2810417H13Rik NM_026515.2 p15(PAF), Pclaf RIKEN cDNA 2810417H13 gene 2900026A02Rik NM_172884.3 Gm449, LOC231620 RIKEN cDNA 2900026A02 gene Abcc8 NM_011510.3 SUR1, Sur, D930031B21Rik ATP-binding cassette, sub-family C (CFTR/MRP), member 8 Acad10 NM_028037.4 2410021P16Rik acyl-Coenzyme A dehydrogenase family, member 10 Acly NM_134037.2 A730098H14Rik ATP citrate lyase Acod1 NM_008392.1 Irg1 aconitate decarboxylase 1 Acot11 NM_025590.4 Thea, 2010309H15Rik, 1110020M10Rik,acyl-CoA Them1, thioesterase BFIT1 11 Acot3 NM_134246.3 PTE-Ia, Pte2a acyl-CoA thioesterase 3 Acox1 NM_015729.2 Acyl-CoA oxidase, AOX, D130055E20Rikacyl-Coenzyme A oxidase 1, palmitoyl Adam19 NM_009616.4 Mltnb a disintegrin and metallopeptidase domain 19 (meltrin beta) Adam8 NM_007403.2 CD156a, MS2, E430039A18Rik, CD156a disintegrin and metallopeptidase domain 8 Adamts1 NM_009621.4 ADAM-TS1, ADAMTS-1, METH-1, METH1a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 Adamts12 NM_175501.2 a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 12 Adamts14 NM_001081127.1 Adamts-14, TS14 a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 Adamts17 NM_001033877.4 AU023434 a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 Adamts2 NM_001277305.1 hPCPNI, ADAM-TS2, a disintegrin and ametalloproteinase disintegrin-like and with metallopeptidase thrombospondin -
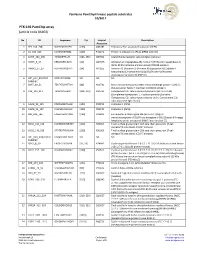
PTK-196 Pamchip Array (Article Code 86402)
PamGene PamChip4 kinase peptide substrates 10/2017 PTK-196 PamChip array (article code 86402) No ID Sequence Tyr Uniprot Description Accession 1 EFS_246_258 GGTDEGIYDVPLL [253] O43281 Embryonal Fyn-associated substrate (HEFS). 2 41_654_666 LDGENIYIRHSNL [660] P11171 Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). 3 ACHD_383_395 YISKAEEYFLLKS [383, 390] Q07001 Acetylcholine receptor subunit delta precursor. 4 AMPE_5_17 EREGSKRYCIQTK [12] Q07075 Glutamyl aminopeptidase (EC 3.4.11.7) (EAP) (Aminopeptidase A) (APA) (Differentiation antigen gp160) (CD249 antigen). 5 ANXA2_17_29 HSTPPSAYGSVKA [24] P07355 Annexin A2 (Annexin-2) (Annexin II) (Lipocortin II) (Calpactin I heavychain) (Chromobindin-8) (p36) (Protein I) (Placental anticoagulantprotein IV) (PAP-IV). 6 ART_004_EAIYAAP EAIYAAPFAKKK NA NA NA FAKKKXC 7 B3AT_39_51 TEATATDYHTTSH [46] P02730 Band 3 anion transport protein (Anion exchange protein 1) (AE 1) (Solute carrier family 4 member 1) (CD233 antigen). 8 C1R_199_211 TEASGYISSLEYP [204, 210] P00736 Complement C1r subcomponent precursor (EC 3.4.21.41) (Complementcomponent 1, r subcomponent) [Contains: Complement C1r subcomponentheavy chain; Complement C1r subcomponent light chain]. 9 CALM_93_105 FDKDGNGYISAAE [100] P0DP23 Calmodulin (CaM). 10 CALM_95_107 KDGNGYISAAELR [100] P0DP23 Calmodulin (CaM). 11 CBL_693_705 EGEEDTEYMTPSS [700] P22681 E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transductionprotein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto-oncogene) (RING finger protein 55). 12 CD3Z_116_128 KDKMAEAYSEIGM [123] P20963 T-cell surface glycoprotein CD3 zeta chain precursor (T-cell receptorT3 zeta chain) (CD247 antigen). 13 CD3Z_146_158 STATKDTYDALHM [153] P20963 T-cell surface glycoprotein CD3 zeta chain precursor (T-cell receptorT3 zeta chain) (CD247 antigen). 14 ART_003_EAI(pY)AAP EAI(pY)AAPFAKKK NA NA NA FAKKKXC 15 CDK2_8_20 EKIGEGTYGVVYK [15, 19] P24941 Cyclin-dependent kinase 2 (EC:2.7.11.22) Cell division protein kinase 2 (EC 2.7.11.22) (p33 protein kinase). -

Aminopeptidases in Cardiovascular and Renal Function. Role As Predictive Renal Injury Biomarkers
International Journal of Molecular Sciences Review Aminopeptidases in Cardiovascular and Renal Function. Role as Predictive Renal Injury Biomarkers Félix Vargas 1, Rosemary Wangesteen 2, Isabel Rodríguez-Gómez 1 and Joaquín García-Estañ 3,* 1 Depto. Fisiologia, Fac. Medicina, Universidad de Granada, 18071 Granada, Spain; [email protected] (F.V.); [email protected] (I.R.-G.) 2 Depto. Ciencias de la salud, Universidad de Jaén, 23071 Jaén, Spain; [email protected] 3 Depto. Fisiologia, Fac. Medicina, IMIB, Universidad de Murcia, 30120 Murcia, Spain * Correspondence: [email protected] Received: 23 June 2020; Accepted: 3 August 2020; Published: 5 August 2020 Abstract: Aminopeptidases (APs) are metalloenzymes that hydrolyze peptides and polypeptides by scission of the N-terminus amino acid and that also participate in the intracellular final digestion of proteins. APs play an important role in protein maturation, signal transduction, and cell-cycle control, among other processes. These enzymes are especially relevant in the control of cardiovascular and renal functions. APs participate in the regulation of the systemic and local renin–angiotensin system and also modulate the activity of neuropeptides, kinins, immunomodulatory peptides, and cytokines, even contributing to cholesterol uptake and angiogenesis. This review focuses on the role of four key APs, aspartyl-, alanyl-, glutamyl-, and leucyl-cystinyl-aminopeptidases, in the control of blood pressure (BP) and renal function and on their association with different cardiovascular and renal diseases. In this context, the effects of AP inhibitors are analyzed as therapeutic tools for BP control and renal diseases. Their role as urinary biomarkers of renal injury is also explored. The enzymatic activities of urinary APs, which act as hydrolyzing peptides on the luminal surface of the renal tubule, have emerged as early predictive renal injury biomarkers in both acute and chronic renal nephropathies, including those induced by nephrotoxic agents, obesity, hypertension, or diabetes. -

Regulation of Aminopeptidase a Expression in Cervical Carcinoma: Role of Tumor–Stromal Interaction and Vascular Endothelial Growth Factor
Laboratory Investigation (2004) 84, 639–648 & 2004 USCAP, Inc All rights reserved 0023-6837/04 $25.00 www.laboratoryinvestigation.org Regulation of aminopeptidase A expression in cervical carcinoma: role of tumor–stromal interaction and vascular endothelial growth factor Takayasu Suganuma1, Kazuhiko Ino1, Kiyosumi Shibata1, Seiji Nomura1, Hiroaki Kajiyama1, Fumitaka Kikkawa1, Nobuo Tsuruoka2 and Shigehiko Mizutani1 1Department of Obstetrics and Gynecology, Nagoya University Graduate School of Medicine, Nagoya, Japan and 2Institute for Health Care Science, Suntory Ltd, Osaka, Japan We previously demonstrated that aminopeptidase A (APA), a membrane-bound metallopeptidase degrading bioactive peptides such as angiotensin II (Ang II), is expressed in neoplastic lesions of the uterine cervix, and that its expression is upregulated as the lesion progresses from cervical intraepithelial neoplasms (CIN) toward invasive squamous cell carcinomas (SCC). The present study investigated the regulatory mechanisms involved in APA expression and its potential role in cervical carcinoma. Immunohistochemical staining in high-grade CIN and SCC tissues showed that APA was strongly expressed at the edge of lesions adjacent to cervical stromal cells. Fluorescence-activated cell sorting analysis demonstrated that cell surface APA expression was extremely low in three human SCC cell lines, SiHa, TCS and CaSki, under basal conditions. However, both contact and noncontact cocultures with human cervical fibroblasts resulted in the induction of APA expression in these SCC cells. APA expression was also induced in vivo when TCS cells were subcutaneously inoculated into nude mice. Furthermore, APA expression and enzymatic activity were enhanced by addition of the conditioned medium (CM) from fibroblast culture, but not by heat-treated CM. Among the various cytokines tested, vascular endothelial growth factor (VEGF) significantly increased APA activity, and induction of APA by the fibroblast CM was partly inhibited by anti-VEGF neutralizing antibody.