DNA Barcoding to Assess Diet of Larval Eastern Hellbenders in North Carolina
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Blackstone River Watershed 2008 Benthic Macroinvertebrate Bioassessment
Technical Memorandum CN 325.2 BLACKSTONE RIVER WATERSHED 2008 BENTHIC MACROINVERTEBRATE BIOASSESSMENT Peter Mitchell Division of Watershed Management Watershed Planning Program Worcester, MA January, 2014 Commonwealth of Massachusetts Executive Office of Energy and Environmental Affairs Richard K. Sullivan, Jr., Secretary Department of Environmental Protection Kenneth L. Kimmell, Commissioner Bureau of Resource Protection Bethany A. Card, Assistant Commissioner (This page intentionally left blank) Contents INTRODUCTION.............................................................................................................................................1 METHODS ......................................................................................................................................................1 Macroinvertebrate Sampling - RBPIII..........................................................................................................1 Macroinvertebrate Sample Processing and Data Analysis .........................................................................4 Habitat Assessment.....................................................................................................................................6 RESULTS AND DISCUSSION........................................................................................................................6 SUMMARY....................................................................................................................................................10 LITERATURE -

Research Report110
~ ~ WISCONSIN DEPARTMENT OF NATURAL RESOURCES A Survey of Rare and Endangered Mayflies of Selected RESEARCH Rivers of Wisconsin by Richard A. Lillie REPORT110 Bureau of Research, Monona December 1995 ~ Abstract The mayfly fauna of 25 rivers and streams in Wisconsin were surveyed during 1991-93 to document the temporal and spatial occurrence patterns of two state endangered mayflies, Acantha metropus pecatonica and Anepeorus simplex. Both species are candidates under review for addition to the federal List of Endang ered and Threatened Wildlife. Based on previous records of occur rence in Wisconsin, sampling was conducted during the period May-July using a combination of sampling methods, including dredges, air-lift pumps, kick-nets, and hand-picking of substrates. No specimens of Anepeorus simplex were collected. Three specimens (nymphs or larvae) of Acanthametropus pecatonica were found in the Black River, one nymph was collected from the lower Wisconsin River, and a partial exuviae was collected from the Chippewa River. Homoeoneuria ammophila was recorded from Wisconsin waters for the first time from the Black River and Sugar River. New site distribution records for the following Wiscon sin special concern species include: Macdunnoa persimplex, Metretopus borealis, Paracloeodes minutus, Parameletus chelifer, Pentagenia vittigera, Cercobrachys sp., and Pseudiron centra/is. Collection of many of the aforementioned species from large rivers appears to be dependent upon sampling sand-bottomed substrates at frequent intervals, as several species were relatively abundant during only very short time spans. Most species were associated with sand substrates in water < 2 m deep. Acantha metropus pecatonica and Anepeorus simplex should continue to be listed as endangered for state purposes and receive a biological rarity ranking of critically imperiled (S1 ranking), and both species should be considered as candidates proposed for listing as endangered or threatened as defined by the Endangered Species Act. -

“Two-Tailed” Baetidae of Ohio January 2013
Ohio EPA Larval Key for the “two-tailed” Baetidae of Ohio January 2013 Larval Key for the “two-tailed” Baetidae of Ohio For additional keys and descriptions see: Ide (1937), Provonsha and McCafferty (1982), McCafferty and Waltz (1990), Lugo-Ortiz and McCafferty (1998), McCafferty and Waltz (1998), Wiersema (2000), McCafferty et al. (2005) and McCafferty et al. (2009). 1. Forecoxae with filamentous gill (may be very small), gills usually with dark clouding, cerci without dark band near middle, claws with a smaller second row of teeth. .............................. ............................................................................................................... Heterocloeon (H.) sp. (Two species, H. curiosum (McDunnough) and H. frivolum (McDunnough), are reported from Ohio, however, the larger hind wing pads used by Morihara and McCafferty (1979) to distinguish H. frivolum have not been verified by OEPA.) Figures from Ide, 1937. Figures from Müller-Liebenau, 1974. 1'. Forecoxae without filamentous gill, other characters variable. .............................................. 2 2. Cerci with alternating pale and dark bands down its entire length, body dorsoventrally flattened, gills with a dark clouded area, hind wing pads greatly reduced. ............................... ......................................................................................... Acentrella parvula (McDunnough) Figure from Ide, 1937. Figure from Wiersema, 2000. 2'. Cerci without alternating pale and dark bands, other characters variable. ............................ -

New State Records of Aquatic Insects for Ohio, U.S.A
Volume 121, Number 1, January and February 2010 75 NEW STATE RECORDS OF AQUATIC INSECTS FOR OHIO, U.S.A. (EPHEMEROPTERA, PLECOPTERA, TRICHOPTERA, COLEOPTERA)1 Michael J. Bolton2 ABSTRACT: Biomonitoring of Ohio streams by the Ohio Environmental Protection Agency has found new state records for the Ephemeroptera (mayflies): Baetis brunneicolor McDunnough, Iswaeon anoka (Daggy), Paracloeodes fleeki McCafferty and Lenat, Plauditus cestus (Provonsha and McCafferty), and Rhithrogena manifesta Eaton; the Plecoptera (stoneflies): Pteronarcys cf. biloba Newman; the Trichop- tera (caddisflies): Brachycentrus numerosus (Say) and Psilotreta rufa (Hagen); and the Coleoptera (bee- tles): Gyretes sinuatus LeConte, Dicranopselaphus variegatus Horn, and Microcylloepus pusillus (Le Conte). Additional records are given for the mayfly Paracloeodes minutus (Daggy). KEY WORDS: Ohio, state record, Ephemeroptera, Plecoptera, Trichoptera, Coleoptera The Ohio Environmental Protection Agency conducts biological and water qual- ity studies of Ohio streams to ascertain the condition of the aquatic resource. One component of these studies is an evaluation of the macroinvertebrate communities. As a result of this sampling, species of aquatic insects in the Ephemeroptera (may- flies), Plecoptera (stoneflies), Trichoptera (caddisflies), and Coleoptera (beetles) orders have been collected that have never been reported from Ohio. Randolph and McCafferty (1998) compiled the first state list of mayflies for Ohio. Gaufin (1956) produced a state list of stoneflies for Ohio with additions by Tkac and Foote (1978), Robertson (1979), and Fishbeck (1987). Listing of species distributions by state in Stewart and Stark (2002) and Stark and Armitage (2000, 2004) incorporated Ohio records found in the various revisionary publications. Huryn and Foote (1983) pro- duced the first comprehensive state list of caddisflies which was amended by Mac Lean and MacLean (1984), Usis and MacLean (1986), Garono and MacLean (1988), Usis and Foote (1989), and Keiper and Bartolotta (2003). -

Waterbody: Northeast Black Creek Basin: Bird Sink
Waterbody: Northeast Black Creek Background Healthy, well-balanced stream communities may be maintained with some level of human activity, but ex- cessive human disturbance may result in waterbody degradation. Human stressors may include increased inputs of nutrients, sediments, and/or other contam- inants from watershed runoff, adverse hydrologic al- terations, undesirable removal of habitat or riparian buffer vegetation, and introduction of exotic plants and animals. Water quality standards are designed to protect designated uses of the waters of the state (e.g., recreation, aquatic life, fish consumption), and exceedances of these standards are associated with interference of the designated use. Basin: Bird Sink Due to ongoing beaver activity, station BC1 is no Northeast Black Creek is a tannic, acidic, predomi- longer sampled. Leon County staff continue to nantly nitrogen-limited stream located in northeast- evaluate the hydrological and plant community ern Leon County. The stream forms near Centerville changes that are occurring in this section. Road and the Chemonie Plantation subdivision and Methods flows southeast through the Miccosukee Land Coop- erative before crossing under Capitola Road. The Surface water samples were collected to determine creek then turns northeast to join Still Creek and then the health of Northeast Black Creek and met the re- flows into Bird Sink. quirements of the Florida Department of Environ- mental Protection (FDEP). As shown in the following pie chart, approximately 31% of the 15,783-acre watershed is comprised of Results urban, agriculture/rangeland, transportation and utilities land uses. Increases in stormwater runoff and Nutrients waterbody nutrient loads can often be attributed to According to FDEP requirements, four temporally in- these types of land uses. -

Ohio EPA Macroinvertebrate Taxonomic Level December 2019 1 Table 1. Current Taxonomic Keys and the Level of Taxonomy Routinely U
Ohio EPA Macroinvertebrate Taxonomic Level December 2019 Table 1. Current taxonomic keys and the level of taxonomy routinely used by the Ohio EPA in streams and rivers for various macroinvertebrate taxonomic classifications. Genera that are reasonably considered to be monotypic in Ohio are also listed. Taxon Subtaxon Taxonomic Level Taxonomic Key(ies) Species Pennak 1989, Thorp & Rogers 2016 Porifera If no gemmules are present identify to family (Spongillidae). Genus Thorp & Rogers 2016 Cnidaria monotypic genera: Cordylophora caspia and Craspedacusta sowerbii Platyhelminthes Class (Turbellaria) Thorp & Rogers 2016 Nemertea Phylum (Nemertea) Thorp & Rogers 2016 Phylum (Nematomorpha) Thorp & Rogers 2016 Nematomorpha Paragordius varius monotypic genus Thorp & Rogers 2016 Genus Thorp & Rogers 2016 Ectoprocta monotypic genera: Cristatella mucedo, Hyalinella punctata, Lophopodella carteri, Paludicella articulata, Pectinatella magnifica, Pottsiella erecta Entoprocta Urnatella gracilis monotypic genus Thorp & Rogers 2016 Polychaeta Class (Polychaeta) Thorp & Rogers 2016 Annelida Oligochaeta Subclass (Oligochaeta) Thorp & Rogers 2016 Hirudinida Species Klemm 1982, Klemm et al. 2015 Anostraca Species Thorp & Rogers 2016 Species (Lynceus Laevicaudata Thorp & Rogers 2016 brachyurus) Spinicaudata Genus Thorp & Rogers 2016 Williams 1972, Thorp & Rogers Isopoda Genus 2016 Holsinger 1972, Thorp & Rogers Amphipoda Genus 2016 Gammaridae: Gammarus Species Holsinger 1972 Crustacea monotypic genera: Apocorophium lacustre, Echinogammarus ischnus, Synurella dentata Species (Taphromysis Mysida Thorp & Rogers 2016 louisianae) Crocker & Barr 1968; Jezerinac 1993, 1995; Jezerinac & Thoma 1984; Taylor 2000; Thoma et al. Cambaridae Species 2005; Thoma & Stocker 2009; Crandall & De Grave 2017; Glon et al. 2018 Species (Palaemon Pennak 1989, Palaemonidae kadiakensis) Thorp & Rogers 2016 1 Ohio EPA Macroinvertebrate Taxonomic Level December 2019 Taxon Subtaxon Taxonomic Level Taxonomic Key(ies) Informal grouping of the Arachnida Hydrachnidia Smith 2001 water mites Genus Morse et al. -

CHAPTER 4: EPHEMEROPTERA (Mayflies)
Guide to Aquatic Invertebrate Families of Mongolia | 2009 CHAPTER 4 EPHEMEROPTERA (Mayflies) EPHEMEROPTERA Draft June 17, 2009 Chapter 4 | EPHEMEROPTERA 45 Guide to Aquatic Invertebrate Families of Mongolia | 2009 ORDER EPHEMEROPTERA Mayflies 4 Mayfly larvae are found in a variety of locations including lakes, wetlands, streams, and rivers, but they are most common and diverse in lotic habitats. They are common and abundant in stream riffles and pools, at lake margins and in some cases lake bottoms. All mayfly larvae are aquatic with terrestrial adults. In most mayfly species the adult only lives for 1-2 days. Consequently, the majority of a mayfly’s life is spent in the water as a larva. The adult lifespan is so short there is no need for the insect to feed and therefore the adult does not possess functional mouthparts. Mayflies are often an indicator of good water quality because most mayflies are relatively intolerant of pollution. Mayflies are also an important food source for fish. Ephemeroptera Morphology Most mayflies have three caudal filaments (tails) (Figure 4.1) although in some taxa the terminal filament (middle tail) is greatly reduced and there appear to be only two caudal filaments (only one genus actually lacks the terminal filament). Mayflies have gills on the dorsal surface of the abdomen (Figure 4.1), but the number and shape of these gills vary widely between taxa. All mayflies possess only one tarsal claw at the end of each leg (Figure 4.1). Characters such as gill shape, gill position, and tarsal claw shape are used to separate different mayfly families. -

Plecoptera: Perlidae), with an Annotated Checklist of the Subfamily in the Realm
Opusc. Zool. Budapest, 2016, 47(2): 173–196 On the identity of some Oriental Acroneuriinae taxa (Plecoptera: Perlidae), with an annotated checklist of the subfamily in the realm D. MURÁNYI1 & W.H. LI2 1Dávid Murányi, Department of Civil and Environmental Engineering, Ehime University, Bunkyo-cho 3, Matsuyama, 790-8577 Japan, and Department of Zoology, Hungarian Natural History Museum, H-1088 Budapest, Baross u. 13, Hungary. E-mail: [email protected], [email protected] 2Weihai Li, Department of Plant Protection, Henan Institute of Science and Technology, Xinxiang, Henan, 453003 China. E-mail: [email protected] Abstract. The monotypic Taiwanese genus Mesoperla Klapálek, 1913 is redescribed on the basis of a male syntype specimen, and its affinities are re-evaluated. The single female type specimen of further two Oriental monotypic genera, Kalidasia Klapálek, 1914 and Nirvania Klapálek, 1914, are confirmed to be lost or destroyed respectively; both genera are considered as nomina dubia. The Sichuan endemic Acroneuria grahami Wu & Claassen, 1934 is redescribed on the basis of male holotype. Distinctive characters of the genus Brahmana Klapálek, 1914 consisting of five, inadequately known Oriental species are discussed. Flavoperla needhami (Klapálek, 1916) and Sinacroneuria sinica (Yang & Yang, 1998) comb. novae are suggested for an Indian species originally described in Gibosia Okamoto, 1912 and a Chinese species originally described in Acroneuria Pictet, 1841. At present, 62 species of Acroneuriinae, classified in 10 valid genera are reported from the Oriental Realm but 29 species are inadequately known. A key is presented to distinguish males of the Asian Acroneuriinae genera. Asian distribution of each genera are detailed and depicted on a map. -

Invertebrate Prey Selectivity of Channel Catfish (Ictalurus Punctatus) in Western South Dakota Prairie Streams Erin D
South Dakota State University Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange Electronic Theses and Dissertations 2017 Invertebrate Prey Selectivity of Channel Catfish (Ictalurus punctatus) in Western South Dakota Prairie Streams Erin D. Peterson South Dakota State University Follow this and additional works at: https://openprairie.sdstate.edu/etd Part of the Aquaculture and Fisheries Commons, and the Terrestrial and Aquatic Ecology Commons Recommended Citation Peterson, Erin D., "Invertebrate Prey Selectivity of Channel Catfish (Ictalurus punctatus) in Western South Dakota Prairie Streams" (2017). Electronic Theses and Dissertations. 1677. https://openprairie.sdstate.edu/etd/1677 This Thesis - Open Access is brought to you for free and open access by Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Open PRAIRIE: Open Public Research Access Institutional Repository and Information Exchange. For more information, please contact [email protected]. INVERTEBRATE PREY SELECTIVITY OF CHANNEL CATFISH (ICTALURUS PUNCTATUS) IN WESTERN SOUTH DAKOTA PRAIRIE STREAMS BY ERIN D. PETERSON A thesis submitted in partial fulfillment of the degree for the Master of Science Major in Wildlife and Fisheries Sciences South Dakota State University 2017 iii ACKNOWLEDGEMENTS South Dakota Game, Fish & Parks provided funding for this project. Oak Lake Field Station and the Department of Natural Resource Management at South Dakota State University provided lab space. My sincerest thanks to my advisor, Dr. Nels H. Troelstrup, Jr., for all of the guidance and support he has provided over the past three years and for taking a chance on me. -

Taxonomic and Distributional Notes on Perlesta Teaysia, P
Grubbs, S.A. & R.E. DeWalt 2008. Taxonomic and distributional notes on Perlesta teaysia, P. golconda, and P. shawnee Plecoptera: Perlidae). Illiesia, 4(14):143-149. Available online: http://www2.pms-lj.si/illiesia/Illiesia04-14.pdf TAXONOMIC AND DISTRIBUTIONAL NOTES ON PERLESTA TEAYSIA, P. GOLCONDA, AND P. SHAWNEE (PLECOPTERA: PERLIDAE) Scott A. Grubbs1 & R. Edward DeWalt2 1 Department of Biology and Center for Biodiversity Studies Western Kentucky University Bowling Green, Kentucky, U.S.A. 42101 E-mail: [email protected] 2 Illinois Natural History Survey 1816 S. Oak Street Champaign, IL 61820 USA E-mail: [email protected] ABSTRACT Perlesta napacola DeWalt is shown to be a junior synonym of P. teaysia Kirchner & Kondratieff. The egg of P. golconda DeWalt & Stark is illustrated with scanning electron microscopy for the first time, and a significant range extension is presented and discussed for P. shawnee Grubbs. Keywords: Plecoptera, Perlidae, Perlesta, P. teaysia, P. napacola, P. golconda, P. shawnee INTRODUCTION Ontario (CNC), C. P. Gillette Museum of Arthropod The known diversity of the eastern Nearctic Diversity, Colorado State University, Fort Collins, stonefly genus Perlesta continues to increase, as Colorado, (CSU), Illinois Natural History Survey, evidenced by the number of new species descriptions Champaign, Illinois (INHS), Ohio Biological Survey, within the last 20 years. Stark (1989) recognized 12 Columbus, Ohio (OBS), Purdue University, West species as distinct. New descriptions published prior Lafayette, Indiana (PU), and Western Kentucky to Stark’s (2004) updated taxonomic treatment, plus University, Bowling Green, Kentucky (WKU). those presented in Kondratieff et al. (2006, 2008), increased the total number of species known to 27 Perlesta teaysia Kirchner & Kondratieff (Stark et al., 2008). -
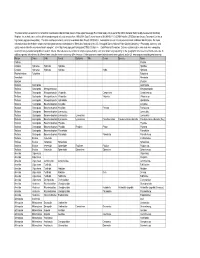
This Table Contains a Taxonomic List of Benthic Invertebrates Collected from Streams in the Upper Mississippi River Basin Study
This table contains a taxonomic list of benthic invertebrates collected from streams in the Upper Mississippi River Basin study unit as part of the USGS National Water Quality Assessemnt (NAWQA) Program. Invertebrates were collected from woody snags in selected streams from 1996-2004. Data Retreival occurred 26-JAN-06 11.10.25 AM from the USGS data warehouse (Taxonomic List Invert http://water.usgs.gov/nawqa/data). The data warehouse currently contains invertebrate data through 09/30/2002. Invertebrate taxa can include provisional and conditional identifications. For more information about invertebrate sample processing and taxonomic standards see, "Methods of analysis by the U.S. Geological Survey National Water Quality Laboratory -- Processing, taxonomy, and quality control of benthic macroinvertebrate samples", at << http://nwql.usgs.gov/Public/pubs/OFR00-212.html >>. Data Retrieval Precaution: Extreme caution must be exercised when comparing taxonomic lists generated using different search criteria. This is because the number of samples represented by each taxa list will vary depending on the geographic criteria selected for the retrievals. In addition, species lists retrieved at different times using the same criteria may differ because: (1) the taxonomic nomenclature (names) were updated, and/or (2) new samples containing new taxa may Phylum Class Order Family Subfamily Tribe Genus Species Taxon Porifera Porifera Cnidaria Hydrozoa Hydroida Hydridae Hydridae Cnidaria Hydrozoa Hydroida Hydridae Hydra Hydra sp. Platyhelminthes Turbellaria Turbellaria Nematoda Nematoda Bryozoa Bryozoa Mollusca Gastropoda Gastropoda Mollusca Gastropoda Mesogastropoda Mesogastropoda Mollusca Gastropoda Mesogastropoda Viviparidae Campeloma Campeloma sp. Mollusca Gastropoda Mesogastropoda Viviparidae Viviparus Viviparus sp. Mollusca Gastropoda Mesogastropoda Hydrobiidae Hydrobiidae Mollusca Gastropoda Basommatophora Ancylidae Ancylidae Mollusca Gastropoda Basommatophora Ancylidae Ferrissia Ferrissia sp. -

Annual Newsletter and Bibliography of the International Society of Plecopterologists
PERLA Annual Newsletter and Bibliography of The International Society of Plecopterologists Capnia valhalla Nelson & Baumann (Capniidae), ♂. California: San Diego Co. Palomar Mountain, Fry Creek. Photograph by C. R. Nelson PERLA NO. 30, 2012 Department of Bioagricultural Sciences and Pest Management Colorado State University Fort Collins, Colorado 80523 USA PERLA Annual Newsletter and Bibliography of the International Society of Plecopterologists Available on Request to the Managing Editor MANAGING EDITOR: Boris C. Kondratieff Department of Bioagricultural Sciences And Pest Management Colorado State University Fort Collins, Colorado 80523 USA E-mail: [email protected] EDITORIAL BOARD: Richard W. Baumann Department of Biology and Monte L. Bean Life Science Museum Brigham Young University Provo, Utah 84602 USA E-mail: [email protected] J. Manuel Tierno de Figueroa Dpto. de Biología Animal Facultad de Ciencias Universidad de Granada 18071 Granada, SPAIN E-mail: [email protected] Kenneth W. Stewart Department of Biological Sciences University of North Texas Denton, Texas 76203, USA E-mail: [email protected] Shigekazu Uchida Aichi Institute of Technology 1247 Yagusa Toyota 470-0392, JAPAN E-mail: [email protected] Peter Zwick Schwarzer Stock 9 D-36110 Schlitz, GERMANY E-mail: [email protected] 2 TABLE OF CONTENTS Subscription policy………………………………………………………..…………….4 2012 XIIIth International Conference on Ephemeroptera, XVIIth International Symposium on Plecoptera in JAPAN…………………………………………………………………………………...5 How to host