SUMO1 Modification of Histone H4 Is Involved in the Pathogenesis of Nodular Lymphocyte Predominant Hodgkin Lymphoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
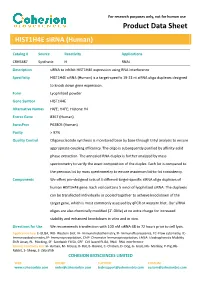
Product Data Sheet
For research purposes only, not for human use Product Data Sheet HIST1H4E siRNA (Human) Catalog # Source Reactivity Applications CRH5487 Synthetic H RNAi Description siRNA to inhibit HIST1H4E expression using RNA interference Specificity HIST1H4E siRNA (Human) is a target-specific 19-23 nt siRNA oligo duplexes designed to knock down gene expression. Form Lyophilized powder Gene Symbol HIST1H4E Alternative Names H4/E; H4FE; Histone H4 Entrez Gene 8367 (Human) SwissProt P62805 (Human) Purity > 97% Quality Control Oligonucleotide synthesis is monitored base by base through trityl analysis to ensure appropriate coupling efficiency. The oligo is subsequently purified by affinity-solid phase extraction. The annealed RNA duplex is further analyzed by mass spectrometry to verify the exact composition of the duplex. Each lot is compared to the previous lot by mass spectrometry to ensure maximum lot-to-lot consistency. Components We offers pre-designed sets of 3 different target-specific siRNA oligo duplexes of human HIST1H4E gene. Each vial contains 5 nmol of lyophilized siRNA. The duplexes can be transfected individually or pooled together to achieve knockdown of the target gene, which is most commonly assessed by qPCR or western blot. Our siRNA oligos are also chemically modified (2’-OMe) at no extra charge for increased stability and enhanced knockdown in vitro and in vivo. Directions for Use We recommends transfection with 100 nM siRNA 48 to 72 hours prior to cell lysis. Application key: E- ELISA, WB- Western blot, IH- Immunohistochemistry, -

Histone Isoform H2A1H Promotes Attainment of Distinct Physiological
Bhattacharya et al. Epigenetics & Chromatin (2017) 10:48 DOI 10.1186/s13072-017-0155-z Epigenetics & Chromatin RESEARCH Open Access Histone isoform H2A1H promotes attainment of distinct physiological states by altering chromatin dynamics Saikat Bhattacharya1,4,6, Divya Reddy1,4, Vinod Jani5†, Nikhil Gadewal3†, Sanket Shah1,4, Raja Reddy2,4, Kakoli Bose2,4, Uddhavesh Sonavane5, Rajendra Joshi5 and Sanjay Gupta1,4* Abstract Background: The distinct functional efects of the replication-dependent histone H2A isoforms have been dem- onstrated; however, the mechanistic basis of the non-redundancy remains unclear. Here, we have investigated the specifc functional contribution of the histone H2A isoform H2A1H, which difers from another isoform H2A2A3 in the identity of only three amino acids. Results: H2A1H exhibits varied expression levels in diferent normal tissues and human cancer cell lines (H2A1C in humans). It also promotes cell proliferation in a context-dependent manner when exogenously overexpressed. To uncover the molecular basis of the non-redundancy, equilibrium unfolding of recombinant H2A1H-H2B dimer was performed. We found that the M51L alteration at the H2A–H2B dimer interface decreases the temperature of melting of H2A1H-H2B by ~ 3 °C as compared to the H2A2A3-H2B dimer. This diference in the dimer stability is also refected in the chromatin dynamics as H2A1H-containing nucleosomes are more stable owing to M51L and K99R substitu- tions. Molecular dynamic simulations suggest that these substitutions increase the number of hydrogen bonds and hydrophobic interactions of H2A1H, enabling it to form more stable nucleosomes. Conclusion: We show that the M51L and K99R substitutions, besides altering the stability of histone–histone and histone–DNA complexes, have the most prominent efect on cell proliferation, suggesting that the nucleosome sta- bility is intimately linked with the physiological efects observed. -

University of California, San Diego
UNIVERSITY OF CALIFORNIA, SAN DIEGO The post-terminal differentiation fate of RNAs revealed by next-generation sequencing A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Biomedical Sciences by Gloria Kuo Lefkowitz Committee in Charge: Professor Benjamin D. Yu, Chair Professor Richard Gallo Professor Bruce A. Hamilton Professor Miles F. Wilkinson Professor Eugene Yeo 2012 Copyright Gloria Kuo Lefkowitz, 2012 All rights reserved. The Dissertation of Gloria Kuo Lefkowitz is approved, and it is acceptable in quality and form for publication on microfilm and electronically: __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ __________________________________________________________________ Chair University of California, San Diego 2012 iii DEDICATION Ma and Ba, for your early indulgence and support. Matt and James, for choosing more practical callings. Roy, my love, for patiently sharing the ups and downs of this journey. iv EPIGRAPH It is foolish to tear one's hair in grief, as though sorrow would be made less by baldness. ~Cicero v TABLE OF CONTENTS Signature Page .............................................................................................................. iii Dedication .................................................................................................................... -

SUMO-1 Regulates the Conformational Dynamics of Thymine-DNA
Smet-Nocca et al. BMC Biochemistry 2011, 12:4 http://www.biomedcentral.com/1471-2091/12/4 RESEARCHARTICLE Open Access SUMO-1 regulates the conformational dynamics of Thymine-DNA Glycosylase regulatory domain and competes with its DNA binding activity Caroline Smet-Nocca1, Jean-Michel Wieruszeski2, Hélène Léger1,3, Sebastian Eilebrecht1,3, Arndt Benecke1,3* Abstract Background: The human thymine-DNA glycosylase (TDG) plays a dual role in base excision repair of G:U/T mismatches and in transcription. Regulation of TDG activity by SUMO-1 conjugation was shown to act on both functions. Furthermore, TDG can interact with SUMO-1 in a non-covalent manner. Results: Using NMR spectroscopy we have determined distinct conformational changes in TDG upon either covalent sumoylation on lysine 330 or intermolecular SUMO-1 binding through a unique SUMO-binding motif (SBM) localized in the C-terminal region of TDG. The non-covalent SUMO-1 binding induces a conformational change of the TDG amino-terminal regulatory domain (RD). Such conformational dynamics do not exist with covalent SUMO-1 attachment and could potentially play a broader role in the regulation of TDG functions for instance during transcription. Both covalent and non-covalent processes activate TDG G:U repair similarly. Surprisingly, despite a dissociation of the SBM/SUMO-1 complex in presence of a DNA substrate, SUMO-1 preserves its ability to stimulate TDG activity indicating that the non-covalent interactions are not directly involved in the regulation of TDG activity. SUMO-1 instead acts, as demonstrated here, indirectly by competing with the regulatory domain of TDG for DNA binding. -

Genome-Wide Approach to Identify Risk Factors for Therapy-Related Myeloid Leukemia
Leukemia (2006) 20, 239–246 & 2006 Nature Publishing Group All rights reserved 0887-6924/06 $30.00 www.nature.com/leu ORIGINAL ARTICLE Genome-wide approach to identify risk factors for therapy-related myeloid leukemia A Bogni1, C Cheng2, W Liu2, W Yang1, J Pfeffer1, S Mukatira3, D French1, JR Downing4, C-H Pui4,5,6 and MV Relling1,6 1Department of Pharmaceutical Sciences, The University of Tennessee, Memphis, TN, USA; 2Department of Biostatistics, The University of Tennessee, Memphis, TN, USA; 3Hartwell Center, The University of Tennessee, Memphis, TN, USA; 4Department of Pathology, The University of Tennessee, Memphis, TN, USA; 5Department of Hematology/Oncology St Jude Children’s Research Hospital, The University of Tennessee, Memphis, TN, USA; and 6Colleges of Medicine and Pharmacy, The University of Tennessee, Memphis, TN, USA Using a target gene approach, only a few host genetic risk therapy increases, the importance of identifying host factors for factors for treatment-related myeloid leukemia (t-ML) have been secondary neoplasms increases. defined. Gene expression microarrays allow for a more 4 genome-wide approach to assess possible genetic risk factors Because DNA microarrays interrogate multiple ( 10 000) for t-ML. We assessed gene expression profiles (n ¼ 12 625 genes in one experiment, they allow for a ‘genome-wide’ probe sets) in diagnostic acute lymphoblastic leukemic cells assessment of genes that may predispose to leukemogenesis. from 228 children treated on protocols that included leukemo- DNA microarray analysis of gene expression has been used to genic agents such as etoposide, 13 of whom developed t-ML. identify distinct expression profiles that are characteristic of Expression of 68 probes, corresponding to 63 genes, was different leukemia subtypes.13,14 Studies using this method have significantly related to risk of t-ML. -

Differences for Ectopic Versus Eutopic Cells
556 RBMO VOLUME 39 ISSUE 4 2019 ARTICLE Chemosensitivity and chemoresistance in endometriosis – differences for ectopic versus eutopic cells BIOGRAPHY Andres Salumets is Professor of Reproductive Medicine at the University of Tartu, and Scientific Head at the Competence Centre on Health Technologies, Tartu, Estonia. He has been involved in assisted reproduction for 20 years, first as an embryologist and later as a researcher. His major interests are endometriosis, endometrial biology and implantation. Darja Lavogina1,2,*, Külli Samuel1, Arina Lavrits1,3, Alvin Meltsov1, Deniss Sõritsa1,4,5, Ülle Kadastik6, Maire Peters1,4, Ago Rinken2, Andres Salumets1,4,7, 8 KEY MESSAGE Akt/PKB inhibitor GSK690693, CK2 inhibitor ARC-775, MAPK pathway inhibitor sorafenib, proteasome inhibitor bortezomib, and microtubule-depolymerizing toxin MMAE showed higher cytotoxicity in eutopic cells. In contrast, 10 µmol/l of the anthracycline toxin doxorubicin caused cellular death in ectopic cells more effectively than in eutopic cells, underlining the potential of doxorubicin in endometriosis research. ABSTRACT Research question: Endometriosis is a common gynaecological disease defined by the presence of endometrium-like tissue outside the uterus. This complex disease, often accompanied by severe pain and infertility, causes a significant medical and socioeconomic burden; hence, novel strategies are being sought for the treatment of endometriosis. Here, we set out to explore the cytotoxic effects of a panel of compounds to find toxins with different efficiency in eutopic versus ectopic cells, thus highlighting alterations in the corresponding molecular pathways. Design: The effect on cellular viability of 14 compounds was established in a cohort of paired eutopic and ectopic endometrial stromal cell samples from 11 patients. -

Cancer TNT Ashwin Ram 12/5/2017 Background: Chromatin Writers, Readers, Erasers
Cancer TNT Ashwin Ram 12/5/2017 Background: Chromatin Writers, Readers, Erasers writer effector eg. HAT, HMT reader eg. bromodomain eraser eg. HDAC, KDM The writer HAT1: A known H4 lysine 5,12 di-acetyltransferase writer siHAT1 siControl HAT1 H4 K12Ac H4 K5Ac actin Western blot for histone H4 modifications after control and HAT1 siRNA transfections. HAT1: EGF-stimulated immunoprecipitation specific specific - - HAT1 IgG HAT1 IgG - - R α non R α non EGF: + + + - - - WB: HAT1 Immunoprecipitation / WB to measure HAT1 levels +/- Heatmap of gene expression changes of all human histone EGF acetyltransferases +/- EGF and siRNA treatments shows HAT1 expression is EGF-dependent Working model of HAT1 : The oldest “new” histone acetyltransferase EGF EGFR plasma membrane HAT1 H4 H3 Rbap46/48 a nuclear membrane H4 H2A H3 H2B HAT1 S phase Surprise: HAT1 also binds (a few sites) on chromatin HAT1 ChipSeq signal sits on Hist1 locus on Chromosome 6 Read Depth HAT1 bound sites (zoom) HAT1 ChIP-seq peaks cluster at Read Depth histone H4 promoters. Hist1H2BE Hist1H4D Hist1H3D Hist1H4E Is HAT1 a transcription factor for its substrate (H4)? EGF EGFR plasma membrane HAT1 H4 H3 Rbap46/48 a nuclear membrane H4 H2A H3 H2B HAT1 S phase HAT1 is required for S-phase burst of histone H4 mRNA HAT1 Hist1H4B mRNA level 50 Rbap46 45 shCont-3 H4 40 shHAT1-A7 35 shHAT1-B6 30 25 B6 A7 - - 20 15 shHAT1 shHAT1 shControl 10 Gene actin)Expression (versus 5 HAT1 0 0 2 4 6 8 10 actin hours after release from double thymidine block G1 S G2/M HAT1 loss: Life with less histones EGF -

Reconstructing Cell Cycle Pseudo Time-Series Via Single-Cell Transcriptome Data—Supplement
School of Natural Sciences and Mathematics Reconstructing Cell Cycle Pseudo Time-Series Via Single-Cell Transcriptome Data—Supplement UT Dallas Author(s): Michael Q. Zhang Rights: CC BY 4.0 (Attribution) ©2017 The Authors Citation: Liu, Zehua, Huazhe Lou, Kaikun Xie, Hao Wang, et al. 2017. "Reconstructing cell cycle pseudo time-series via single-cell transcriptome data." Nature Communications 8, doi:10.1038/s41467-017-00039-z This document is being made freely available by the Eugene McDermott Library of the University of Texas at Dallas with permission of the copyright owner. All rights are reserved under United States copyright law unless specified otherwise. File name: Supplementary Information Description: Supplementary figures, supplementary tables, supplementary notes, supplementary methods and supplementary references. CCNE1 CCNE1 CCNE1 CCNE1 36 40 32 34 32 35 30 32 28 30 30 28 28 26 24 25 Normalized Expression Normalized Expression Normalized Expression Normalized Expression 26 G1 S G2/M G1 S G2/M G1 S G2/M G1 S G2/M Cell Cycle Stage Cell Cycle Stage Cell Cycle Stage Cell Cycle Stage CCNE1 CCNE1 CCNE1 CCNE1 40 32 40 40 35 30 38 30 30 28 36 25 26 20 20 34 Normalized Expression Normalized Expression Normalized Expression 24 Normalized Expression G1 S G2/M G1 S G2/M G1 S G2/M G1 S G2/M Cell Cycle Stage Cell Cycle Stage Cell Cycle Stage Cell Cycle Stage Supplementary Figure 1 | High stochasticity of single-cell gene expression means, as demonstrated by relative expression levels of gene Ccne1 using the mESC-SMARTer data. For every panel, 20 sample cells were randomly selected for each of the three stages, followed by plotting the mean expression levels at each stage. -

T-Cell Receptor (TCR) Signaling Promotes the Assembly of Ranbp2
RESEARCH ARTICLE T-cell receptor (TCR) signaling promotes the assembly of RanBP2/RanGAP1- SUMO1/Ubc9 nuclear pore subcomplex via PKC--mediated phosphorylation of RanGAP1 Yujiao He1, Zhiguo Yang1†, Chen-si Zhao1†, Zhihui Xiao1†, Yu Gong1, Yun-Yi Li1, Yiqi Chen1, Yunting Du1, Dianying Feng1, Amnon Altman2, Yingqiu Li1* 1MOE Key Laboratory of Gene Function and Regulation, Guangdong Province Key Laboratory of Pharmaceutical Functional Genes, State Key Laboratory of Biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China; 2Center for Cancer Immunotherapy, La Jolla Institute for Immunology, La Jolla, United States Abstract The nuclear pore complex (NPC) is the sole and selective gateway for nuclear transport, and its dysfunction has been associated with many diseases. The metazoan NPC subcomplex RanBP2, which consists of RanBP2 (Nup358), RanGAP1-SUMO1, and Ubc9, regulates the assembly and function of the NPC. The roles of immune signaling in regulation of NPC remain poorly understood. Here, we show that in human and murine T cells, following T-cell receptor (TCR) stimulation, protein kinase C-q (PKC-q) directly phosphorylates RanGAP1 to facilitate RanBP2 subcomplex assembly and nuclear import and, thus, the nuclear translocation of AP-1 transcription *For correspondence: factor. Mechanistically, TCR stimulation induces the translocation of activated PKC-q to the NPC, 504 506 [email protected] where it interacts with and phosphorylates RanGAP1 on Ser and Ser . RanGAP1 phosphorylation increases its binding affinity for Ubc9, thereby promoting sumoylation of RanGAP1 †These authors contributed and, finally, assembly of the RanBP2 subcomplex. Our findings reveal an unexpected role of PKC-q equally to this work as a direct regulator of nuclear import and uncover a phosphorylation-dependent sumoylation of Competing interests: The RanGAP1, delineating a novel link between TCR signaling and assembly of the RanBP2 NPC authors declare that no subcomplex. -

(12) United States Patent (10) Patent No.: US 7,799,528 B2 Civin Et Al
US007799528B2 (12) United States Patent (10) Patent No.: US 7,799,528 B2 Civin et al. (45) Date of Patent: Sep. 21, 2010 (54) THERAPEUTIC AND DIAGNOSTIC Al-Hajj et al., “Prospective identification of tumorigenic breast can APPLICATIONS OF GENES cer cells.” Proc. Natl. Acad. Sci. U.S.A., 100(7):3983-3988 (2003). DIFFERENTIALLY EXPRESSED IN Bhatia et al., “A newly discovered class of human hematopoietic cells LYMPHO-HEMATOPOETC STEM CELLS with SCID-repopulating activity.” Nat. Med. 4(9)1038-45 (1998). (75) Inventors: Curt I. Civin, Baltimore, MD (US); Bonnet, D., “Normal and leukemic CD35-negative human Robert W. Georgantas, III, Towson, hematopoletic stem cells.” Rev. Clin. Exp. Hematol. 5:42-61 (2001). Cambot et al., “Human Immune Associated Nucleotide 1: a member MD (US) of a new guanosine triphosphatase family expressed in resting T and (73) Assignee: The Johns Hopkins University, B cells.” Blood, 99(9):3293-3301 (2002). Baltimore, MD (US) Chen et al., “Kruppel-like Factor 4 (Gut-enriched Kruppel-like Fac tor) Inhibits Cell Proliferation by Blocking G1/S Progression of the *) NotOt1Ce: Subjubject to anyy d1Sclaimer,disclai theh term off thisthi Cell Cycle.” J. Biol. Chem., 276(32):30423-30428 (2001). patent is extended or adjusted under 35 Chen et al., “Transcriptional profiling of Kurppel-like factor 4 reveals U.S.C. 154(b) by 168 days. a function in cell cycle regulation and epithelial differentiation.” J. Mol. Biol. 326(3):665-677 (2003). (21) Appl. No.: 11/199.665 Civin et al., “Highly purified CD34-positive cells reconstitute (22) Filed: Aug. 9, 2005 hematopoiesis,” J. -

The E3 Ligase PIAS1 Regulates P53 Sumoylation to Control Stress-Induced Apoptosis of Lens
fcell-09-660494 June 14, 2021 Time: 13:5 # 1 ORIGINAL RESEARCH published: 14 June 2021 doi: 10.3389/fcell.2021.660494 The E3 Ligase PIAS1 Regulates p53 Sumoylation to Control Stress-Induced Apoptosis of Lens Edited by: Wolfgang Knabe, Epithelial Cells Through the Universität Münster, Germany Proapoptotic Regulator Bax Reviewed by: Guillaume Bossis, † † † † † Centre National de la Recherche Qian Nie , Huimin Chen , Ming Zou , Ling Wang , Min Hou , Jia-Wen Xiang, Scientifique (CNRS), France Zhongwen Luo, Xiao-Dong Gong, Jia-Ling Fu, Yan Wang, Shu-Yu Zheng, Yuan Xiao, Stefan Müller, Yu-Wen Gan, Qian Gao, Yue-Yue Bai, Jing-Miao Wang, Lan Zhang, Xiang-Cheng Tang, Goethe University Frankfurt, Germany Xuebin Hu, Lili Gong, Yizhi Liu* and David Wan-Cheng Li*‡ Leena Latonen, University of Eastern Finland, Finland State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-sen University, Guangzhou, China *Correspondence: David Wan-Cheng Li Protein sumoylation is one of the most important post-translational modifications [email protected] Yizhi Liu regulating many biological processes (Flotho A & Melchior F. 2013. Ann Rev. Biochem. [email protected] 82:357–85). Our previous studies have shown that sumoylation plays a fundamental †These authors have contributed role in regulating lens differentiation (Yan et al., 2010. PNAS, 107(49):21034-9.; equally to this work Gong et al., 2014. PNAS. 111(15):5574–9). Whether sumoylation is implicated in lens ‡ ORCID: David Wan-Cheng Li pathogenesis remains elusive. Here, we present evidence to show that the protein orcid.org/0000-0002-7398-7630 inhibitor of activated STAT-1 (PIAS1), a E3 ligase for sumoylation, is implicated in regulating stress-induced lens pathogenesis. -

Histone-Related Genes Are Hypermethylated in Lung Cancer
Published OnlineFirst October 1, 2019; DOI: 10.1158/0008-5472.CAN-19-1019 Cancer Genome and Epigenome Research Histone-Related Genes Are Hypermethylated in Lung Cancer and Hypermethylated HIST1H4F Could Serve as a Pan-Cancer Biomarker Shihua Dong1,Wei Li1, Lin Wang2, Jie Hu3,Yuanlin Song3, Baolong Zhang1, Xiaoguang Ren1, Shimeng Ji3, Jin Li1, Peng Xu1, Ying Liang1, Gang Chen4, Jia-Tao Lou2, and Wenqiang Yu1 Abstract Lung cancer is the leading cause of cancer-related deaths lated in all 17 tumor types from TCGA datasets (n ¼ 7,344), worldwide. Cytologic examination is the current "gold stan- which was further validated in nine different types of cancer dard" for lung cancer diagnosis, however, this has low sensi- (n ¼ 243). These results demonstrate that HIST1H4F can tivity. Here, we identified a typical methylation signature of function as a universal-cancer-only methylation (UCOM) histone genes in lung cancer by whole-genome DNA methyl- marker, which may aid in understanding general tumorigen- ation analysis, which was validated by The Cancer Genome esis and improve screening for early cancer diagnosis. Atlas (TCGA) lung cancer cohort (n ¼ 907) and was further confirmed in 265 bronchoalveolar lavage fluid samples with Significance: These findings identify a new biomarker for specificity and sensitivity of 96.7% and 87.0%, respectively. cancer detection and show that hypermethylation of histone- More importantly, HIST1H4F was universally hypermethy- related genes seems to persist across cancers. Introduction to its low specificity, LDCT is far from satisfactory as a screening tool for clinical application, similar to other currently used cancer Lung cancer is one of the most common malignant tumors and biomarkers, such as carcinoembryonic antigen (CEA), neuron- the leading cause of cancer-related deaths worldwide (1, 2).