Ancestors Aplacophoran Molluscs and Their Derivation from Chiton-Like a Molecular Palaeobiological Hypothesis for the Origin Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Some Aspects of the Biology of Three Northwestern Atlantic Chitons
University of New Hampshire University of New Hampshire Scholars' Repository Doctoral Dissertations Student Scholarship Spring 1978 SOME ASPECTS OF THE BIOLOGY OF THREE NORTHWESTERN ATLANTIC CHITONS: TONICELLA RUBRA, TONICELLA MARMOREA, AND ISCHNOCHITON ALBUS (MOLLUSCA: POLYPLACOPHORA) PAUL DAVID LANGER University of New Hampshire, Durham Follow this and additional works at: https://scholars.unh.edu/dissertation Recommended Citation LANGER, PAUL DAVID, "SOME ASPECTS OF THE BIOLOGY OF THREE NORTHWESTERN ATLANTIC CHITONS: TONICELLA RUBRA, TONICELLA MARMOREA, AND ISCHNOCHITON ALBUS (MOLLUSCA: POLYPLACOPHORA)" (1978). Doctoral Dissertations. 2329. https://scholars.unh.edu/dissertation/2329 This Dissertation is brought to you for free and open access by the Student Scholarship at University of New Hampshire Scholars' Repository. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of University of New Hampshire Scholars' Repository. For more information, please contact [email protected]. INFORMATION TO USERS This material was produced from a microfilm copy of the original document. While the most advanced technological means to photograph and reproduce this document have been used, the quality is heavily dependent upon the quality of the original submitted. The following explanation of techniques is provided to help you understand markings or patterns which may appear on this reproduction. 1.The sign or "target" for pages apparently lacking from the document photographed is "Missing Page(s)". If it was possible to obtain the missing page(s) or section, they are spliced into the film along with adjacent pages. This may have necessitated cutting thru an image and duplicating adjacent pages to insure you complete continuity. 2. When an image on the film is obliterated with a large round black mark, it is an indication that the photographer suspected that the copy may have moved during exposure and thus cause a blurred image. -
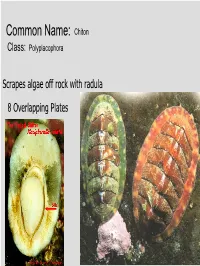
Common Name: Chiton Class: Polyplacophora
Common Name: Chiton Class: Polyplacophora Scrapes algae off rock with radula 8 Overlapping Plates Phylum? Mollusca Class? Gastropoda Common name? Brown sea hare Class? Scaphopoda Common name? Tooth shell or tusk shell Mud Tentacle Foot Class? Gastropoda Common name? Limpet Phylum? Mollusca Class? Bivalvia Class? Gastropoda Common name? Brown sea hare Phylum? Mollusca Class? Gastropoda Common name? Nudibranch Class? Cephalopoda Cuttlefish Octopus Squid Nautilus Phylum? Mollusca Class? Gastropoda Most Bivalves are Filter Feeders A B E D C • A: Mantle • B: Gill • C: Mantle • D: Foot • E: Posterior adductor muscle I.D. Green: Foot I.D. Red Gills Three Body Regions 1. Head – Foot 2. Visceral Mass 3. Mantle A B C D • A: Radula • B: Mantle • C: Mouth • D: Foot What are these? Snail Radulas Dorsal HingeA Growth line UmboB (Anterior) Ventral ByssalC threads Mussel – View of Outer Shell • A: Hinge • B: Umbo • C: Byssal threads Internal Anatomy of the Bay Mussel A B C D • A: Labial palps • B: Mantle • C: Foot • D: Byssal threads NacreousB layer Posterior adductorC PeriostracumA muscle SiphonD Mantle Byssal threads E Internal Anatomy of the Bay Mussel • A: Periostracum • B: Nacreous layer • C: Posterior adductor muscle • D: Siphon • E: Mantle Byssal gland Mantle Gill Foot Labial palp Mantle Byssal threads Gill Byssal gland Mantle Foot Incurrent siphon Byssal Labial palp threads C D B A E • A: Foot • B: Gills • C: Posterior adductor muscle • D: Excurrent siphon • E: Incurrent siphon Heart G F H E D A B C • A: Foot • B: Gills • C: Mantle • D: Excurrent siphon • E: Incurrent siphon • F: Posterior adductor muscle • G: Labial palps • H: Anterior adductor muscle Siphon or 1. -

Marine Invertebrate Field Guide
Marine Invertebrate Field Guide Contents ANEMONES ....................................................................................................................................................................................... 2 AGGREGATING ANEMONE (ANTHOPLEURA ELEGANTISSIMA) ............................................................................................................................... 2 BROODING ANEMONE (EPIACTIS PROLIFERA) ................................................................................................................................................... 2 CHRISTMAS ANEMONE (URTICINA CRASSICORNIS) ............................................................................................................................................ 3 PLUMOSE ANEMONE (METRIDIUM SENILE) ..................................................................................................................................................... 3 BARNACLES ....................................................................................................................................................................................... 4 ACORN BARNACLE (BALANUS GLANDULA) ....................................................................................................................................................... 4 HAYSTACK BARNACLE (SEMIBALANUS CARIOSUS) .............................................................................................................................................. 4 CHITONS ........................................................................................................................................................................................... -

Chitons (Mollusca: Polyplacophora) Known from Benthic Monitoring Programs in the Southern California Bight
ISSN 0738-9388 THE FESTIVUS A publication of the San Diego Shell Club Volume XLI Special Issue June 11, 2009 Chitons (Mollusca: Polyplacophora) Known from Benthic Monitoring Programs in the Southern California Bight Timothy D. Stebbins and Douglas J. Eernisse COVER PHOTO Live specimen of Lepidozona sp. C occurring on a piece of metal debris collected off San Diego, southern California at a depth of 90 m. Photo provided courtesy of R. Rowe. Vol. XLI(6): 2009 THE FESTIVUS Page 53 CHITONS (MOLLUSCA: POLYPLACOPHORA) KNOWN FROM BENTHIC MONITORING PROGRAMS IN THE SOUTHERN CALIFORNIA BIGHT TIMOTHY D. STEBBINS 1,* and DOUGLAS J. EERNISSE 2 1 City of San Diego Marine Biology Laboratory, Metropolitan Wastewater Department, San Diego, CA, USA 2 Department of Biological Science, California State University, Fullerton, CA, USA Abstract: About 36 species of chitons possibly occur at depths greater than 30 m along the continental shelf and slope of the Southern California Bight (SCB), although little is known about their distribution or ecology. Nineteen species are reported here based on chitons collected as part of long-term, local benthic monitoring programs or less frequent region-wide surveys of the entire SCB, and these show little overlap with species that occur at depths typically encountered by scuba divers. Most chitons were collected between 30-305 m depths, although records are included for a few from slightly shallower waters. Of the two extant chiton lineages, Lepidopleurida is represented by Leptochitonidae (2 genera, 3 species), while Chitonida is represented by Ischnochitonidae (2 genera, 6-9 species) and Mopaliidae (4 genera, 7 species). -

Constraints on the Timescale of Animal Evolutionary History
Palaeontologia Electronica palaeo-electronica.org Constraints on the timescale of animal evolutionary history Michael J. Benton, Philip C.J. Donoghue, Robert J. Asher, Matt Friedman, Thomas J. Near, and Jakob Vinther ABSTRACT Dating the tree of life is a core endeavor in evolutionary biology. Rates of evolution are fundamental to nearly every evolutionary model and process. Rates need dates. There is much debate on the most appropriate and reasonable ways in which to date the tree of life, and recent work has highlighted some confusions and complexities that can be avoided. Whether phylogenetic trees are dated after they have been estab- lished, or as part of the process of tree finding, practitioners need to know which cali- brations to use. We emphasize the importance of identifying crown (not stem) fossils, levels of confidence in their attribution to the crown, current chronostratigraphic preci- sion, the primacy of the host geological formation and asymmetric confidence intervals. Here we present calibrations for 88 key nodes across the phylogeny of animals, rang- ing from the root of Metazoa to the last common ancestor of Homo sapiens. Close attention to detail is constantly required: for example, the classic bird-mammal date (base of crown Amniota) has often been given as 310-315 Ma; the 2014 international time scale indicates a minimum age of 318 Ma. Michael J. Benton. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Philip C.J. Donoghue. School of Earth Sciences, University of Bristol, Bristol, BS8 1RJ, U.K. [email protected] Robert J. -

Download Preprint
1 Mobilising molluscan models and genomes in biology 2 Angus Davison1 and Maurine Neiman2 3 1. School of Life Sciences, University Park, University of Nottingham, NG7 2RD, UK 4 2. Department of Biology, University of Iowa, Iowa City, IA, USA and Department of Gender, 5 Women's, and Sexuality Studies, University of Iowa, Iowa, City, IA, USA 6 Abstract 7 Molluscs are amongst the most ancient, diverse, and important of all animal taxa. Even so, 8 no individual mollusc species has emerged as a broadly applied model system in biology. 9 We here make the case that both perceptual and methodological barriers have played a role 10 in the relative neglect of molluscs as research organisms. We then summarize the current 11 application and potential of molluscs and their genomes to address important questions in 12 animal biology, and the state of the field when it comes to the availability of resources such 13 as genome assemblies, cell lines, and other key elements necessary to mobilising the 14 development of molluscan model systems. We conclude by contending that a cohesive 15 research community that works together to elevate multiple molluscan systems to ‘model’ 16 status will create new opportunities in addressing basic and applied biological problems, 17 including general features of animal evolution. 18 Introduction 19 Molluscs are globally important as sources of food, calcium and pearls, and as vectors of 20 human disease. From an evolutionary perspective, molluscs are notable for their remarkable 21 diversity: originating over 500 million years ago, there are over 70,000 extant mollusc 22 species [1], with molluscs present in virtually every ecosystem. -

Geobiological Events in the Ediacaran Period
Geobiological Events in the Ediacaran Period Shuhai Xiao Department of Geosciences, Virginia Tech, Blacksburg, VA 24061, USA NSF; NASA; PRF; NSFC; Virginia Tech Geobiology Group; CAS; UNLV; UCR; ASU; UMD; Amherst; Subcommission of Neoproterozoic Stratigraphy; 1 Goals To review biological (e.g., acanthomorphic acritarchs; animals; rangeomorphs; biomineralizing animals), chemical (e.g., carbon and sulfur isotopes, oxygenation of deep oceans), and climatic (e.g., glaciations) events in the Ediacaran Period; To discuss integration and future directions in Ediacaran geobiology; 2 Knoll and Walter, 1992 • Acanthomorphic acritarchs in early and Ediacara fauna in late Ediacaran Period; • Strong carbon isotope variations; • Varanger-Laplandian glaciation; • What has happened since 1992? 3 Age Constraints: South China (538.2±1.5 Ma) 541 Ma Cambrian Dengying Ediacaran Sinian 551.1±0.7 Ma Doushantuo 632.5±0.5 Ma 635 Ma 635.2±0.6 Ma Nantuo (Tillite) 636 ± 5Ma Cryogenian Nanhuan 654 ± 4Ma Datangpo 663±4 Ma Neoproterozoic Neoproterozoic Jiangkou Group Banxi Group 725±10 Ma Tonian Qingbaikouan 1000 Ma • South China radiometric ages: Condon et al., 2005; Hoffmann et al., 2004; Zhou et al., 2004; Bowring et al., 2007; S. Zhang et al., 2008; Q. Zhang et al., 2008; • Additional ages from Nama Group (Namibia), Conception Group (Newfoundland), and Vendian (White Sea); 4 The Ediacaran Period Ediacara fossils Cambrian 545 Ma Nama assemblage 555 Ma White Sea assemblage 565 Ma Avalon assemblage 575 Ma 585 Ma Doushantuo biota 595 Ma 605 Ma Ediacaran Period 615 Ma -

Probabilistic Methods Surpass Parsimony When Assessing Clade Support in Phylogenetic Analyses of Discrete Morphological Data
O'Reilly, J., Puttick, M., Pisani, D., & Donoghue, P. (2017). Probabilistic methods surpass parsimony when assessing clade support in phylogenetic analyses of discrete morphological data. Palaeontology. https://doi.org/10.1111/pala.12330 Publisher's PDF, also known as Version of record License (if available): CC BY Link to published version (if available): 10.1111/pala.12330 Link to publication record in Explore Bristol Research PDF-document This is the final published version of the article (version of record). It first appeared online via Wiley at http://onlinelibrary.wiley.com/doi/10.1111/pala.12330/abstract. Please refer to any applicable terms of use of the publisher. University of Bristol - Explore Bristol Research General rights This document is made available in accordance with publisher policies. Please cite only the published version using the reference above. Full terms of use are available: http://www.bristol.ac.uk/red/research-policy/pure/user-guides/ebr-terms/ [Palaeontology, 2017, pp. 1–14] PROBABILISTIC METHODS SURPASS PARSIMONY WHEN ASSESSING CLADE SUPPORT IN PHYLOGENETIC ANALYSES OF DISCRETE MORPHOLOGICAL DATA by JOSEPH E. O’REILLY1 ,MARKN.PUTTICK1,2 , DAVIDE PISANI1,3 and PHILIP C. J. DONOGHUE1 1School of Earth Sciences, University of Bristol, Life Sciences Building, Tyndall Avenue, Bristol, BS8 1TQ, UK; [email protected]; [email protected]; [email protected]; [email protected] 2Department of Earth Sciences, The Natural History Museum, Cromwell Road, London, SW7 5BD, UK 3School of Biological Sciences, University of Bristol, Life Sciences Building, Tyndall Avenue, Bristol, BS8 1TQ, UK Typescript received 28 April 2017; accepted in revised form 13 September 2017 Abstract: Fossil taxa are critical to inferences of historical 50% support. -

Contributions in BIOLOGY and GEOLOGY
MILWAUKEE PUBLIC MUSEUM Contributions In BIOLOGY and GEOLOGY Number 51 November 29, 1982 A Compendium of Fossil Marine Families J. John Sepkoski, Jr. MILWAUKEE PUBLIC MUSEUM Contributions in BIOLOGY and GEOLOGY Number 51 November 29, 1982 A COMPENDIUM OF FOSSIL MARINE FAMILIES J. JOHN SEPKOSKI, JR. Department of the Geophysical Sciences University of Chicago REVIEWERS FOR THIS PUBLICATION: Robert Gernant, University of Wisconsin-Milwaukee David M. Raup, Field Museum of Natural History Frederick R. Schram, San Diego Natural History Museum Peter M. Sheehan, Milwaukee Public Museum ISBN 0-893260-081-9 Milwaukee Public Museum Press Published by the Order of the Board of Trustees CONTENTS Abstract ---- ---------- -- - ----------------------- 2 Introduction -- --- -- ------ - - - ------- - ----------- - - - 2 Compendium ----------------------------- -- ------ 6 Protozoa ----- - ------- - - - -- -- - -------- - ------ - 6 Porifera------------- --- ---------------------- 9 Archaeocyatha -- - ------ - ------ - - -- ---------- - - - - 14 Coelenterata -- - -- --- -- - - -- - - - - -- - -- - -- - - -- -- - -- 17 Platyhelminthes - - -- - - - -- - - -- - -- - -- - -- -- --- - - - - - - 24 Rhynchocoela - ---- - - - - ---- --- ---- - - ----------- - 24 Priapulida ------ ---- - - - - -- - - -- - ------ - -- ------ 24 Nematoda - -- - --- --- -- - -- --- - -- --- ---- -- - - -- -- 24 Mollusca ------------- --- --------------- ------ 24 Sipunculida ---------- --- ------------ ---- -- --- - 46 Echiurida ------ - --- - - - - - --- --- - -- --- - -- - - --- -

Can Molecular Clocks and the Fossil Record Be Reconciled?
Prospects & Overviews Review essays The origin of animals: Can molecular clocks and the fossil record be reconciled? John A. Cunningham1)2)Ã, Alexander G. Liu1)†, Stefan Bengtson2) and Philip C. J. Donoghue1) The evolutionary emergence of animals is one of the most Introduction significant episodes in the history of life, but its timing remains poorly constrained. Molecular clocks estimate that The apparent absence of a fossil record prior to the appearance of trilobites in the Cambrian famously troubled Darwin. He animals originated and began diversifying over 100 million wrote in On the origin of species that if his theory of evolution years before the first definitive metazoan fossil evidence in were true “it is indisputable that before the lowest [Cambrian] the Cambrian. However, closer inspection reveals that clock stratum was deposited ... the world swarmed with living estimates and the fossil record are less divergent than is creatures.” Furthermore, he could give “no satisfactory answer” often claimed. Modern clock analyses do not predict the as to why older fossiliferous deposits had not been found [1]. In the intervening century and a half, a record of Precambrian presence of the crown-representatives of most animal phyla fossils has been discovered extending back over three billion in the Neoproterozoic. Furthermore, despite challenges years (popularly summarized in [2]). Nevertheless, “Darwin’s provided by incomplete preservation, a paucity of phylo- dilemma” regarding the origin and early evolution of Metazoa genetically informative characters, and uncertain expecta- arguably persists, because incontrovertible fossil evidence for tions of the anatomy of early animals, a number of animals remains largely, or some might say completely, absent Neoproterozoic fossils can reasonably be interpreted as from Neoproterozoic rocks [3]. -

Recent Advances and Unanswered Questions in Deep Molluscan Phylogenetics Author(S): Kevin M
Recent Advances and Unanswered Questions in Deep Molluscan Phylogenetics Author(s): Kevin M. Kocot Source: American Malacological Bulletin, 31(1):195-208. 2013. Published By: American Malacological Society DOI: http://dx.doi.org/10.4003/006.031.0112 URL: http://www.bioone.org/doi/full/10.4003/006.031.0112 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Amer. Malac. Bull. 31(1): 195–208 (2013) Recent advances and unanswered questions in deep molluscan phylogenetics* Kevin M. Kocot Auburn University, Department of Biological Sciences, 101 Rouse Life Sciences, Auburn University, Auburn, Alabama 36849, U.S.A. Correspondence, Kevin M. Kocot: [email protected] Abstract. Despite the diversity and importance of Mollusca, evolutionary relationships among the eight major lineages have been a longstanding unanswered question in Malacology. Early molecular studies of deep molluscan phylogeny, largely based on nuclear ribosomal gene data, as well as morphological cladistic analyses largely failed to provide robust hypotheses of relationships among major lineages. -

Aspects of the Ecology of a Littoral Chiton, Sypharochiton Pellisekpentis (Mollusca: Polyplacophora)
New Zealand Journal of Marine and Freshwater Research ISSN: 0028-8330 (Print) 1175-8805 (Online) Journal homepage: http://www.tandfonline.com/loi/tnzm20 Aspects of the ecology of a littoral chiton, Sypharochiton pellisekpentis (Mollusca: Polyplacophora) P. R. Boyle To cite this article: P. R. Boyle (1970) Aspects of the ecology of a littoral chiton, Sypharochiton pellisekpentis (Mollusca: Polyplacophora), New Zealand Journal of Marine and Freshwater Research, 4:4, 364-384, DOI: 10.1080/00288330.1970.9515354 To link to this article: http://dx.doi.org/10.1080/00288330.1970.9515354 Published online: 30 Mar 2010. Submit your article to this journal Article views: 263 View related articles Citing articles: 13 View citing articles Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=tnzm20 Download by: [203.118.161.175] Date: 14 February 2017, At: 22:55 364 [DEC. ASPECTS OF THE ECOLOGY OF A LITTORAL CHITON, SYPHAROCHITON PELLISEKPENTIS (MOLLUSCA: POLYPLACOPHORA) P. R. BOYLE* Department of Zoology, University of Auckland (Received for publication 23 May 1969) SUMMARY On several Auckland shores, a littoral chiton, Sypharochiton pelliserpentis (Quoy and Gaimard, 1835), was widely distributed and common. At Castor Bay it was the commonest chiton, and its density equalled or exceeded that of the commonest limpet {Cellana spp.) over most of the inter-tidal range. Spot measurements of population density were made at other sites including exposed and sheltered shores. The smallest animals were restricted to the lower shore in pools or on areas df rock which were slow to drain. Exclusive of these small animals, the population structure was similar in pools and water- filled crevices situated either high or low on the shore.