Genetic Analysis of Citron (Citrus Medica L.) Using Simple Sequence
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Diversity and Population Structure of Pummelo (Citrus Maxima) Germplasm in China
Tree Genetics & Genomes (2017) 13: 58 DOI 10.1007/s11295-017-1133-0 ORIGINAL ARTICLE Genetic diversity and population structure of pummelo (Citrus maxima) germplasm in China Huiwen Yu1 & Xiaoming Yang 1 & Fei Guo1 & Xiaolin Jiang1 & Xiuxin Deng1 & Qiang Xu1 Received: 31 July 2016 /Revised: 11 March 2017 /Accepted: 19 March 2017 /Published online: 26 April 2017 # Springer-Verlag Berlin Heidelberg 2017 Abstract Pummelo (Citrus maxima) is one of the basic spe- Keywords Pummelo . Genetic diversity . Population cies of Citrus. It has been cultivated for about 4000 years in structure . Nuclear simple sequence repeat (nSSR) China, and therefore, there are abundant germplasm during the long time of culture. However, there is still a lack of a detailed study of the genetic characteristics of pummelo pop- Introduction ulation. In this study, genetic diversity and population struc- ture among 274 pummelo accessions collected in China were Citrus is one of the most important fruit crops in the world. analyzed using 31 nuclear simple sequence repeat (nSSR) The genetic background of citrus is very complicated because markers. The observed heterozygosity was calculated as of its biological characteristics, such as wide sexual compati- 0.325 and genetic differentiation Fst as 0.077. Genetic struc- bility on interspecies and intergenus levels. The complex ge- ture analysis divided the whole germplasm into three subpop- netic background has hindered the genetic studies in citrus. ulations, Pop-a, Pop-b, and Pop-c. Pop-a was composed of Exploring genetic variation within a single species will facil- accessions mostly from Southeast China, Pop-b was com- itate genetic analysis such as genome-wide association studies posed of accessions from the central region of South China, (GWAS) of important traits. -

Texas Citrus Tree Valuesjuan R
EHT-011 7/13 Texas Citrus Tree ValuesJuan R. Anciso and Luis A. Ribera* Photo courtesy of Rod Santa Ana ommercial citrus growers and homeowners To figure the total value per tree for years 2 to often need to determine the value of their 6, add the value of a tree the previous year plus mature citrus trees. The objective is to deter- the potential gross revenue the new tree is bring- mine the value of a mature grapefruit tree (Rio ing. RedC variety) and a mature orange tree (Valencia Finally, calculate the total value of mature variety) to assess the value of a tree or an orchard. trees (7 years and older) by adding the cumula- Grapefruit and orange trees usually reach full tive potential net revenue of a mature tree over 7 maturity by year 7 with an estimated yield of 23 years, plus the planting cost minus the cumulative and 18 tons per acre, respectively (Table 1.) How- potential net revenue of a new tree, years 1 to 6. ever, some production is expected from year 2 Overall, citrus tree values vary from year 1 with yearly increments until they reach maturity. through 7. At year 7, citrus trees are fully mature The price per ton used in this study is the due to their commercial production potential. Of 5-year average (2008–2012) producers received course, the longevity or lifespan of a citrus tree for either grapefruit or Valencia oranges. The cost depends on its care and whether it endures any of production is separated into planting costs or type of cold injury and rehabilitation, but the esti- establishment costs and annual production cost. -

A History of Fruits on the Southeast Asian Mainland
OFFPRINT A history of fruits on the Southeast Asian mainland Roger Blench Kay Williamson Educational Foundation Cambridge, UK E-mail: [email protected] http://www.rogerblench.info/RBOP.htm Occasional Paper 4 Linguistics, Archaeology and the Human Past Edited by Toshiki OSADA and Akinori UESUGI Indus Project Research Institute for Humanity and Nature, Kyoto, Japan 2008 ISBN 978-4-902325-33-1 A history of Fruits on the Southeast Asian mainland A history of fruits on the Southeast Asian mainland Roger Blench Kay Williamson Educational Foundation Cambridge, UK E-mail: [email protected] http://www.rogerblench.info/RBOP.htm ABSTRACT The paper presents an overview of the history of the principal tree fruits grown on the Southeast Asian mainland, making use of data from biogeography, archaeobotany, iconography and linguistics. Many assertions in the literature about the origins of particular species are found to be without empirical basis. In the absence of other data, comparative linguistics is an important source for tracing the spread of some fruits. Contrary to the Pacific, it seems that many of the fruits we now consider characteristic of the region may well have spread in recent times. INTRODUCTION empirical base for Pacific languages is not matched for mainland phyla such as Austroasiatic, Daic, Sino- This study 1) is intended to complement a previous Tibetan or Hmong-Mien, so accounts based purely paper on the history of tree-fruits in island Southeast on Austronesian tend to give a one-sided picture. Asia and the Pacific (Blench 2005). Arboriculture Although occasional detailed accounts of individual is very neglected in comparison to other types of languages exist (e.g. -

Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid
Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid Diaphorina Liberibacter citri Plant Name asiaticus Citrus Huanglongbing Psyllid Aegle marmelos (L.) Corr. Serr.: bael, Bengal quince, golden apple, bela, milva X Aeglopsis chevalieri Swingle: Chevalier’s aeglopsis X X Afraegle gabonensis (Swingle) Engl.: Gabon powder-flask X Afraegle paniculata (Schum.) Engl.: Nigerian powder- flask X Atalantia missionis (Wall. ex Wight) Oliv.: see Pamburus missionis X X Atalantia monophylla (L.) Corr.: Indian atalantia X Balsamocitrus dawei Stapf: Uganda powder- flask X X Burkillanthus malaccensis (Ridl.) Swingle: Malay ghost-lime X Calodendrum capense Thunb.: Cape chestnut X × Citroncirus webberi J. Ingram & H. E. Moore: citrange X Citropsis gilletiana Swingle & M. Kellerman: Gillet’s cherry-orange X Citropsis schweinfurthii (Engl.) Swingle & Kellerm.: African cherry- orange X Citrus amblycarpa (Hassk.) Ochse: djerook leemo, djeruk-limau X Citrus aurantiifolia (Christm.) Swingle: lime, Key lime, Persian lime, lima, limón agrio, limón ceutí, lima mejicana, limero X X Citrus aurantium L.: sour orange, Seville orange, bigarde, marmalade orange, naranja agria, naranja amarga X Citrus depressa Hayata: shiikuwasha, shekwasha, sequasse X Citrus grandis (L.) Osbeck: see Citrus maxima X Citrus hassaku hort. ex Tanaka: hassaku orange X Citrus hystrix DC.: Mauritius papeda, Kaffir lime X X Citrus ichangensis Swingle: Ichang papeda X Citrus jambhiri Lushington: rough lemon, jambhiri-orange, limón rugoso, rugoso X X Citrus junos Sieb. ex Tanaka: xiang -

Tropical Horticulture: Lecture 32 1
Tropical Horticulture: Lecture 32 Lecture 32 Citrus Citrus: Citrus spp., Rutaceae Citrus are subtropical, evergreen plants originating in southeast Asia and the Malay archipelago but the precise origins are obscure. There are about 1600 species in the subfamily Aurantioideae. The tribe Citreae has 13 genera, most of which are graft and cross compatible with the genus Citrus. There are some tropical species (pomelo). All Citrus combined are the most important fruit crop next to grape. 1 Tropical Horticulture: Lecture 32 The common features are a superior ovary on a raised disc, transparent (pellucid) dots on leaves, and the presence of aromatic oils in leaves and fruits. Citrus has increased in importance in the United States with the development of frozen concentrate which is much superior to canned citrus juice. Per-capita consumption in the US is extremely high. Citrus mitis (calamondin), a miniature orange, is widely grown as an ornamental house pot plant. History Citrus is first mentioned in Chinese literature in 2200 BCE. First citrus in Europe seems to have been the citron, a fruit which has religious significance in Jewish festivals. Mentioned in 310 BCE by Theophrastus. Lemons and limes and sour orange may have been mutations of the citron. The Romans grew sour orange and lemons in 50–100 CE; the first mention of sweet orange in Europe was made in 1400. Columbus brought citrus on his second voyage in 1493 and the first plantation started in Haiti. In 1565 the first citrus was brought to the US in Saint Augustine. 2 Tropical Horticulture: Lecture 32 Taxonomy Citrus classification based on morphology of mature fruit (e.g. -

Classification and Cultivars
1 Classification and Cultivars 2 Two Tribes • Clauseneae • Citreae has 3 Subtribes –Triphasiinae –Balsamocitrineae –Citrinae 3 Fortunella • Four species - Small trees and shrubs. • Flowers later than Citrus. • Freeze - hardy • Small fruit –‘Meiwa’ and ‘Marumi’ - round –‘Nagami’ ovate 4 Poncirus • Two trifoliate spp. –trifoliata ‘Flying Dragon’ –poyandra • Deciduous • Thorny, Cold hardy, long thorns • Makes great hedges , rootstocks 5 Microcitrus • Northeastern rainforest Australia • Moderate-sized trees. • Leaves are unifoliate dimorphic • Microcitrus australasica –Resistant to burrowing nematode and phytophthora • Micro leaves, flowers, and fruit 6 Clymenia • Unifoliate acuminate leaves tapering into very short petiole. • Branches are thornless. • Style shorter than other true Citrus and stigma is larger and flattened • Fruit - ovoid, thin peeled, many oil glands, many small seeds. 7 Eremocitrus • Xerophytic native of Australia • Spreading long drooping branches • Leaves unifoliate, greyish green, thick, leatherly, and lanceolate. • Sunken stomata, freeze hardy • Ideal xeroscape plant. 8 Citrus - Subgenus Eucitrus • Vesicles - no acrid or bitter oil • C. medica (Citrons) –Uses - candied peel, • Jewish ceremony • Exocortis indicator 9 Citrus limon (Lemons) • Commerce –‘Lisbon’ and ‘Eureka’ • Dooryard –Meyer (Lemon hybrid) • Rough Lemon –Rootstock 10 Lemon Hybrids • Lemonage (lemon x sweet orange) • Lemonime (lemon x lime) • Lemandrin (lemon x mandarin) • Eremolemon (Eremocitrus x lemon) - Australian Desert Lemon 11 Citrus aurantifolia (Limes) • ‘Key’ or ‘Mexican’ limes • ‘Tahiti’ or ‘Persian’ limes some are triploids and seedless • C. macrophylla (lime-like fruit) –Rootstock in California • Lemonimes (lime x lemon) • Limequats (lime x kumquat) 12 • Not grown either in Tahiti or Persian (Iran) • Seedless and marketed when still dark green 13 C. aurantium - Sour Orange • ‘Seville’ in Southern Europe –Orange marmalade • ‘Bouquet’ & ‘Bergamot’ • - Italy –Essential oil • Many forms like ‘Bittersweet’ –Rootstock - High quality fruit. -

Citrus Trifoliata (Rutaceae): Review of Biology and Distribution in the USA
Nesom, G.L. 2014. Citrus trifoliata (Rutaceae): Review of biology and distribution in the USA. Phytoneuron 2014-46: 1–14. Published 1 May 2014. ISSN 2153 733X CITRUS TRIFOLIATA (RUTACEAE): REVIEW OF BIOLOGY AND DISTRIBUTION IN THE USA GUY L. NESOM 2925 Hartwood Drive Fort Worth, Texas 76109 www.guynesom.com ABSTRACT Citrus trifoliata (aka Poncirus trifoliata , trifoliate orange) has become an aggressive colonizer in the southeastern USA, spreading from plantings as a horticultural novelty and use as a hedge. Its currently known naturalized distribution apparently has resulted from many independent introductions from widely dispersed plantings. Seed set is primarily apomictic and the plants are successful in a variety of habitats, in ruderal habits and disturbed communities as well as in intact natural communities from closed canopy bottomlands to open, upland woods. Trifoliate orange is native to southeastern China and Korea. It was introduced into the USA in the early 1800's but apparently was not widely planted until the late 1800's and early 1900's and was not documented as naturalizing until about 1910. Citrus trifoliata L. (trifoliate orange, hardy orange, Chinese bitter orange, mock orange, winter hardy bitter lemon, Japanese bitter lemon) is a deciduous shrub or small tree relatively common in the southeastern USA. The species is native to eastern Asia and has become naturalized in the USA in many habitats, including ruderal sites as well as intact natural commmunities. It has often been grown as a dense hedge and as a horticultural curiosity because of its green stems and stout green thorns (stipular spines), large, white, fragrant flowers, and often prolific production of persistent, golf-ball sized orange fruits that mature in September and October. -
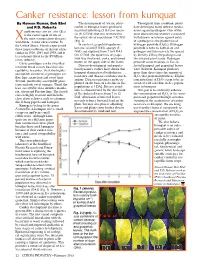
Canker Resistance: Lesson from Kumquat by Naveen Kumar, Bob Ebel the Development of Asiatic Citrus Throughout Their Evolution, Plants and P.D
Canker resistance: lesson from kumquat By Naveen Kumar, Bob Ebel The development of Asiatic citrus Throughout their evolution, plants and P.D. Roberts canker in kumquat leaves produced have developed many defense mecha- anthomonas citri pv. citri (Xcc) localized yellowing (5 DAI) or necro- nisms against pathogens. One of the is the causal agent of one of sis (9-12 DAI) that was restricted to most characteristic features associated the most serious citrus diseases the actual site of inoculation 7-12 DAI with disease resistance against entry X (Fig. 2). of a pathogen is the production of worldwide, Asiatic citrus canker. In the United States, Florida experienced In contrast, grapefruit epidermis hydrogen peroxide (H2O2). Hydrogen three major outbreaks of Asiatic citrus became raised (5 DAI), spongy (5 peroxide is toxic to both plant and canker in 1910, 1984 and 1995, and it DAI) and ruptured from 7 to 8 DAI. pathogen and thus restricts the spread is a constant threat to the $9 billion On 12 DAI, the epidermis of grape- by directly killing the pathogen and citrus industry. fruit was thickened, corky, and turned the infected plant tissue. Hydrogen Citrus genotypes can be classified brown on the upper side of the leaves. peroxide concentrations in Xcc-in- into four broad classes based on sus- Disease development and popula- fected kumquat and grapefruit leaves ceptibility to canker. First, the highly- tion dynamics studies have shown that were different. Kumquat produces susceptible commercial genotypes are kumquat demonstrated both disease more than three times the amount of Key lime, grapefruit and sweet lime. -

PRUNING GUIDELINES Tools: Loppers, Saw, Clippers 10% Bleach and Water Solution
Varieties - choose one that you will want to eat often, as you will have them much of the year (Four Winds Citrus Variety Chart link in your Resources list) - certain citrus mature earlier than others (see early ripening handout) - Unique varieties: - Blood orange: red flesh is antioxidant rich. Often sweeter than other oranges - Yuzu: very little, but very flavorful juice used by chefs. Believed to be a cross between a sour mandarin and ichang papeda - Keiffir lime: regular lime with bumpy skin. Attractive tree with segmented leaves that are extremely fragrant and prized by chefs. - Buddhas hand: not much juice, but very fragrant pith and rind. Odd shaped and often used as an ornamental - Australian finger lime: oblong green lime with many small lime-flavored orbs inside. Called "citrus caviar" Standard varieties of citrus trees often grow to a height of 20 to 30 feet and the canopy -- or width of a tree -- can spread to 18 to 30 feet depending on the variety. Dwarf citrus trees are significantly shorter and narrower, which provides greater flexibility in planting location. Most varieties top out at 8 feet in height with a proportionally smaller canopy. Despite the differences in height and width, regular, semi-dwarf and dwarf citrus varieties produce the same size fruit. -First off DO NOT PRUNE! Those damaged leaves can actually provide protection for the plant until the air warms up. The plant needs to rally and recover. Pruning might just put it over the edge. Sometimes the plants must remain with that ‘raggedy’ appearance until as late as June and in some cases a full year. -

Citrus from Seed?
Which citrus fruits will come true to type Orogrande, Tomatera, Fina, Nour, Hernandina, Clementard.) from seed? Ellendale Tom McClendon writes in Hardy Citrus Encore for the South East: Fortune Fremont (50% monoembryonic) “Most common citrus such as oranges, Temple grapefruit, lemons and most mandarins Ugli Umatilla are polyembryonic and will come true to Wilking type. Because most citrus have this trait, Highly polyembryonic citrus types : will mostly hybridization can be very difficult to produce nucellar polyembryonic seeds that will grow true to type. achieve…. This unique characteristic Citrus × aurantiifolia Mexican lime (Key lime, West allows amateurs to grow citrus from seed, Indian lime) something you can’t do with, say, Citrus × insitorum (×Citroncirus webberii) Citranges, such as Rusk, Troyer etc. apples.” [12*] Citrus × jambhiri ‘Rough lemon’, ‘Rangpur’ lime, ‘Otaheite’ lime Monoembryonic (don’t come true) Citrus × limettioides Palestine lime (Indian sweet lime) Citrus × microcarpa ‘Calamondin’ Meyer Lemon Citrus × paradisi Grapefruit (Marsh, Star Ruby, Nagami Kumquat Redblush, Chironja, Smooth Flat Seville) Marumi Kumquat Citrus × sinensis Sweet oranges (Blonde, navel and Pummelos blood oranges) Temple Tangor Citrus amblycarpa 'Nasnaran' mandarin Clementine Mandarin Citrus depressa ‘Shekwasha’ mandarin Citrus karna ‘Karna’, ‘Khatta’ Poncirus Trifoliata Citrus kinokuni ‘Kishu mandarin’ Citrus lycopersicaeformis ‘Kokni’ or ‘Monkey mandarin’ Polyembryonic (come true) Citrus macrophylla ‘Alemow’ Most Oranges Citrus reshni ‘Cleopatra’ mandarin Changshou Kumquat Citrus sunki (Citrus reticulata var. austera) Sour mandarin Meiwa Kumquat (mostly polyembryonic) Citrus trifoliata (Poncirus trifoliata) Trifoliate orange Most Satsumas and Tangerines The following mandarin varieties are polyembryonic: Most Lemons Dancy Most Limes Emperor Grapefruits Empress Tangelos Fairchild Kinnow Highly monoembryonic citrus types: Mediterranean (Avana, Tardivo di Ciaculli) Will produce zygotic monoembryonic seeds that will not Naartje come true to type. -

UC Riverside UC Riverside Electronic Theses and Dissertations
UC Riverside UC Riverside Electronic Theses and Dissertations Title Cross-Compatibility, Graft-Compatibility, and Phylogenetic Relationships in the Aurantioideae: New Data From the Balsamocitrinae Permalink https://escholarship.org/uc/item/1904r6x3 Author Siebert Wooldridge, Toni Jean Publication Date 2016 Supplemental Material https://escholarship.org/uc/item/1904r6x3#supplemental Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA RIVERSIDE Cross-Compatibility, Graft-Compatibility, and Phylogenetic Relationships in the Aurantioideae: New Data From the Balsamocitrinae A Thesis submitted in partial satisfaction of the requirements for the degree of Master of Science in Plant Biology by Toni J Siebert Wooldridge December 2016 Thesis committee: Dr. Norman C. Ellstrand, Chairperson Dr. Timothy J. Close Dr. Robert R. Krueger The Thesis of Toni J Siebert Wooldridge is approved: Committee Chairperson University of California, Riverside ACKNOWLEDGEMENTS I am indebted to many people who have been an integral part of my research and supportive throughout my graduate studies: A huge thank you to Dr. Norman Ellstrand as my major professor and graduate advisor, and to my supervisor, Dr. Tracy Kahn, who helped influence my decision to go back to graduate school while allowing me to continue my full-time employment with the UC Riverside Citrus Variety Collection. Norm and Tracy, my UCR parents, provided such amazing enthusiasm, guidance and friendship while I was working, going to school and caring for my growing family. Their support was critical and I could not have done this without them. My committee members, Dr. Timothy Close and Dr. Robert Krueger for their valuable advice, feedback and suggestions. -

Citrus Industry Biosecurity Plan 2015
Industry Biosecurity Plan for the Citrus Industry Version 3.0 July 2015 PLANT HEALTH AUSTRALIA | Citrus Industry Biosecurity Plan 2015 Location: Level 1 1 Phipps Close DEAKIN ACT 2600 Phone: +61 2 6215 7700 Fax: +61 2 6260 4321 E-mail: [email protected] Visit our web site: www.planthealthaustralia.com.au An electronic copy of this plan is available through the email address listed above. © Plant Health Australia Limited 2004 Copyright in this publication is owned by Plant Health Australia Limited, except when content has been provided by other contributors, in which case copyright may be owned by another person. With the exception of any material protected by a trade mark, this publication is licensed under a Creative Commons Attribution-No Derivs 3.0 Australia licence. Any use of this publication, other than as authorised under this licence or copyright law, is prohibited. http://creativecommons.org/licenses/by-nd/3.0/ - This details the relevant licence conditions, including the full legal code. This licence allows for redistribution, commercial and non-commercial, as long as it is passed along unchanged and in whole, with credit to Plant Health Australia (as below). In referencing this document, the preferred citation is: Plant Health Australia Ltd (2004) Industry Biosecurity Plan for the Citrus Industry (Version 3.0 – July 2015). Plant Health Australia, Canberra, ACT. Disclaimer: The material contained in this publication is produced for general information only. It is not intended as professional advice on any particular matter. No person should act or fail to act on the basis of any material contained in this publication without first obtaining specific and independent professional advice.