Plant CLE Peptides from Two Distinct Functional Classes Synergistically Induce Division of Vascular Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

PLANT ANATOMY Lecture 15 - Stele and Bundle Types
Jim Bidlack - BIO 5354/4354 PLANT ANATOMY Lecture 15 - Stele and Bundle Types I. Types of steles (stellar types); stele refers to the central vascular network if the shoot and the root (includes outermost phloem and everything to the inside of it) A. Protostele 1. No pith and usually no separate vascular bundles 2. First pattern (phylogenetically) that appeared in vascular plants 3. Predominant pattern found in roots of almost all plants and shoots of lower vascular plants 4. Types of protostele a) Haplostele - xylem is a circular mass b) Actinostele - xylem margin is not smooth; it "undulates" c) Plectostele - xylem not one mass; series of plates B. Siphonostele 1. Has a pith and can have separate vascular bundles (primary growth) 2. More advanced plants 3. Found in shoots of seed plants and in roots having a broad stele (monocots) 4. Types of siphonostele a) Amphiphloic - phloem found on both sides of the xylem 1) Solenostele - very few or little leaf gaps widely separated (continuous cylinder) 2) Dictyostele - leaf gaps are abundant b) Ectophloic - phloem found only to outside of the xylem 1) Eustele (dicots) - one ring of vascular bundles surround a pith 2) Atactostele (monocots) - scattered bundle arrangement II. Stellar patterns at the node (Nodal pattern) A. Different because this is where leaves and buds are attached B. Area above & behind the leaf or bud is the leaf gap C. Area showing where the leaf (bundle(s)) attaches to stem is called leaf trace D. Examples of patterns: 1. Unilacunar, 1 trace; unilacunar, 3 trace; trilacunar, 3 trace III. -

Effect of Stele Type in the Growth Rotation of Dalbergia Melanoxylon
International Journal of Plant and Forestry Sciences Vol. 2, No. 3, May 2015, pp. 1 -10 Available online at http://ijpfs.com/ Research article Effect of Stele type in the growth rotation of Dalbergia melanoxylon. Dr. Washa B. Washa Mkwawa University College of Education Private Bag, MUCE Iringa Tanzania. E-mail: [email protected], +255 752 356 709 This work is licensed under a Creative Commons Attribution 4.0 International License. _____________________________________________________________________________________________ Abstract Examination of the effect of stele type in Dalbergia melanoxylon growth was conducted. Examination of stems to observe stele type and tissue water potential of Barreveld Faux, Cupressus sempervirens and Dalbergia melanoxylon was carried between 2nd – 23rd April 2015 at Mkwawa University Laboratories. About 120 stained stem sections were examined for stele type while other 120 stem pieces were incubated in Nacl solution for water potential. Forty (40) stained sections indicated ectophloic siphonostele in Barreveld Faux and C. sempervirens while 40 sections of D. melanoxylon indicated atactostele. Calculated water potential of Barreveld Faux was -0.01158597 bars and that of C. sempervirens was -0.01257201 bars while that of D. melanoxylon was -0.00320463 bars. Results found that, low water potential in D. melanoxylon is a factor for its slow growth rotation and this is due to the atactostele type existing in D. melanoxylon. A hard Blackwood and valued wood in D. melanoxylon is brought by this stele type. In order to initiate rapid growth rotation in D. melanoxylon is recommended to conduct genetic recombination of the D. melanoxylon with other species which have an easily growing wood although this can lower the hardwood quality and value of the D. -

Stelar Evolution
Stelar System of Plant: Definition and Types Definition of Stelar System: According to the older botanists, the vascular bundle is the fundamental unit in the vascular system of pteridophytes and higher plants. Van Tieghem and Douliot (1886) interpreted the plant body of vascular plant in the different way. According to them, the fundamental parts of a shoot are the cortex and a central cylinder, is known as stele. Thus the stele is defined as a central vascular cylinder, with or without pith and delimited the cortex by endodermis. The term stele has been derived from a Greek word meaning pillar. Van Tieghem and Douliot (1886) recognized only three types of steles. They also thought that the monostelic shoot were rare in comparison of polystelic shoots. It is an established fact that all shoots are monostelic and polystelic condition rarely occurs. The stele of the stem remains connected with that of leaf by a vascular connection known as the leaf supply. Types of Steles: 1. Protostele: Jeffrey (1898), for the first time pointed out the stelar theory from the point of view of the phylogeny. According to him, the primitive type of stele is protostele. In protostele, the vascular tissue is a solid mass and the central core of the xylem is completely surrounded by the strand of phloem. This is the most primitive and simplest type of stele. There are several forms of protostele: (a) Haplostele: This is the most primitive type of protostele. Here the central solid smooth core of xylem remains surrounded by phloem (e.g., in Selaginella spp.). -

The Stele – a Developmental Perspective on the Diversity and Evolution of Primary Vascular 2 Architecture 3 4 Alexandru M.F
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 31 May 2020 doi:10.20944/preprints202005.0472.v1 1 The stele – a developmental perspective on the diversity and evolution of primary vascular 2 architecture 3 4 Alexandru M.F. Tomescu 5 6 Department of Biological Sciences, Humboldt State University, Arcata, California 95521, USA 7 (+1) 707‐826‐3229 [email protected] 8 9 Abstract 10 The stele concept is one of the oldest enduring concepts in plant biology. This paper reviews the 11 concept and its foundations, and builds an argument for an updated view of steles and their evolution. 12 The history of studies of stelar organization has generated a widely ranging array of definitions of the 13 stele that determine the way we classify steles and construct scenarios about the evolution of stelar 14 architecture. Because at the level of the organism biological evolution proceeds by, and is reflected in, 15 changes in development, concepts of structure need to be grounded in development in order to be 16 relevant in an evolutionary perspective. For the stele, most of the traditional definitions that 17 incorporate development have viewed it as the totality of tissues that either originate from procambium 18 – currently the prevailing view – or are bordered by a boundary layer (e.g., endodermis). A definition of 19 the stele that would bring consensus between these perspectives recasts the stele as a structural entity 20 of dual nature. Here, I review briefly the history of the stele concept, basic terminology related to stelar 21 organization, and traditional classifications of the steles. -

An Anatomical Study of the Hawaiian Fern Adenophorus Sarmentosus'
An Anatomical Study of the Hawaiian Fern Adenophorus sarmentosus' KENNETH A. WILSON2 and FRED R. RICKSON:; THE ENDEMIC HAWAIIAN FERN, Adenopborus tative means of propagation is known not only sarm entosus ( Brack.) K. A. Wilson, occurs on in A. sarmentosus, but also in the closely re all of the major islands, growing on moss lated A. haalilioanus, and should be looked for covered trees and occasionally also on mossy in the rare A. pimlatifidus. Similar reproductive rocks. As a representative of the large but poorly behavior has been reported recently in Asple understood fern family Grammitidaceae, A. sat nium plenum of Florida (Wagner, 1963). mentosus was chosen for anatomical studies in The petioles are short, less than 2 em long, order to contribute some information on the crowded on the rhizome, and bearing simple or family which may be of value in later systematic branched deciduous, reddish-brown hairs. The .studies of the group. Anatomical or morpho blades are pinnatifid, elliptic-lanceolate, 8-15 cm logical studies of the grammitids are rare. A long, 1-2 .5 cm wide, and narrowing gradually series of recent papers by Nozu (1958-1960) at both ends, often becoming prolonged into a presents the only anatomical investigation of long caudate apex (Fig. 1, 3) . The venation is members of this family except for a few notes pinnate, with free simple (rarely branched ) published earlier by Ogura (19 38) . veins with clavate or punctiform ends which do The plant material used was collected at W ai not extend to the margins of the blade ( Figs. -

VI. Ferns I: the Marattiales and the Polypodiales, Vegetative Features We Now Take up the Ferns, Two Orders That Together Inclu
VI. Ferns I: The Marattiales and the Polypodiales, Vegetative Features We now take up the ferns, two orders that together include about 12,000 species. Members of these two orders have megaphylls that bear sporangia either abaxially or (rarely) on the margin of the leaf. In addition, they are all spore-dispersed. In this lab, we'll consider the Marattiales, a group of large tropical ferns with primitive features, and the vegetative features of the Polypodiales, the true ferns. A. Marattiales, an Order of Eusporangiate Ferns The Marattiales have a well-documented history. They first appear as tree ferns in the coal swamps right in there with Lepidodendron and Calamites. The living species are prominent in some hot forests, both in tropical America and tropical Asia. They are very like the true ferns (Polypodiales), but they differ in having the common, primitive, thick-walled sporangium, the eusporangium, and in having a distinctive stele and root structure. 1. Living Plants The large ferns on the tables in the lab this week are members of two genera of the Marattiales, Marattia and Angiopteris. a.These plants, like all ferns, have megaphylls. These megaphylls are divided into leaflets called pinnae, which are often divided even further. The feather-like design of these leaves is common among the ferns, suggesting that ferns have some sort of narrow definition to the kinds of leaf design they can evolve. b. The leaflets are borne on stem-like axes called rachises, which, as you can see, have swollen bases on some of the plants in the lab. -

The Phloem Sieve Tube As a Dynamic Carrier of Water and Sugars by Ryan Stanfield a Thesis Submitted in Partial Fulfillment of Th
The Phloem Sieve Tube as a Dynamic Carrier of Water and Sugars by Ryan Stanfield A thesis submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Forest Biology and Management Department of Renewable Resources University of Alberta © Ryan Stanfield, 2019 ABSTRACT This dissertation delves into the anatomy and physiology of the phloem plant vascular tissue. Two primary topics are investigated in this thesis: (1) The cellular localization and response of aquaporin protein water channels in phloem sieve tubes and (2) the mathematical modeling of phloem sieve plates and radial water flows on the hydraulic resistance and pressure profiles of sieve tubes in balsam poplar (Populus balsamifera L.). In the first study, I use immunohistochemistry experiments to identify the distribution and cellular location of aquaporins in the phloem within different organs of the poplar tree. I found that throughout the entire transport pathway from source to sink, sieve tube plasma membranes contain two subfamilies of aquaporin proteins. The first type, PIP1, facilitates the transport of water and some uncharged molecules through the plasma membrane. The second type, PIP2, is the primary water carrying channel in plants. I found that PIP1 aquaporins are mostly found within internal cellular compartments of the sieve tube. This contrasts with PIP2 aquaporins which were found mostly in the plasma membrane. The second study followed the results of the first and looked at how the application of a cold block affected the expression of selected PIP1 and PIP2 genes in phloem tissue. It was found that cold block application transiently increased the abundance of PIP2 aquaporins in the plasma membrane of sieve tubes and altered the abundance of aquaporin mRNA transcript. -

Internal Organisation of Plants
INTERNAL ORGANISATION OF PLANTS • In unicellular and colonial forms every cell behaves as an independent unit and performs all functions • Levels of organisation in plant Anatomy are Tissues - Tissue systems - Organs - Plant body. • Study of tissues and tissue systems of plant body is called Histology • A group of similar or dissimilar cells that have a common origin and function is called tissue. • Tissues are formed as a response to division of labour. Tissues are of two types namely. A) Meristematic tissue B) Permanent tissue. A) Meristamatic tissue: A group of undifferentiated cells having the power of cell division is called Meristematic tissue or Formative tissue. The term 'meristem' was coined by K. Nageli (1858) Characters of meristematic tissue i) Cells are smallin size and isodiametric, cubical or polyhedral in shape. ii) Cells are young and immature iii) Cells are arranged compactly without intercellular spaces. iv) Cell wall is thin and formed of cellulose. v) Dense cytoplasm and abundant with numerous smaller vacuoles vi) Proplastids are present. vii) Ergastic substances absent. viii) Prominent big Nucleus is present ix) Cells divide continously and show active metabolism. Types of Meristems: Basing on origin, meristems are classified into two types namely Primary meristems & Secondary meristems. Meristems which originate from embryonic tissues and continue to remain active in mature parts of the plant is called primary meristems. (mainly seen in apices of root which is continuation of radical) and main stem which is continuation of Plumule) Meristem derived from permanent cells by dedifferentiation is called secondary meristem. Eg: Interfascicular cambium, and cork cambium etc. Secondary meristems are lateral in position parallel to the periphery and help in secondary growth. -
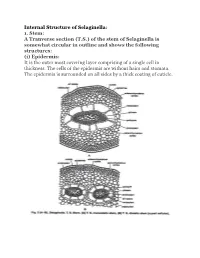
Internal Structure of Selaginella: 1. Stem: a Tranverse Section (TS)
Internal Structure of Selaginella: 1. Stem: A Tranverse section (T.S.) of the stem of Selaginella is somewhat circular in outline and shows the following structures: (i) Epidermis: It is the outer most covering layer comprising of a single cell in thickness. The cells of the epidermis are without hairs and stomata. The epidermis is surrounded on all sides by a thick coating of cuticle. (ii) Cortex: Inner to the epidermis is present a well-defined zone of cortex. The cortex may or may not be differentiated into inner and outer cortex. In case of S. selaginoides, the whole of the cortex is made up of parenchymatous cells while in S. kraussiana, it is differentiated into sclerenchymatous outer cortex and parenchymatous inner cortex. The parenchymatous cortex is usually made up of angular cells i.e., without intercellular spaces but in some cases the cells are rounded and provided with a few inter-cellular spaces. (iii) Stele: The central portion of the stem is occupied by a well-developed stele. The stele is of protostelic type i.e., xylem is present in the centre and surrounded by phloem on all sides. Phloem, in turn, is surrounded by a single layered pericycle. Pith is absent. The stele remains suspended in the centre by radially elongated tubular, unicellular structures known as trabeculae. These are formed by the radial elongation of the endodermal cells. Trabeculae are provided with conspicuous casparian strips. In between the trabeculae are present large spaces known as air spaces. The number of stele is variable in different species of Selaginella. -

Molecular Genetic Framework for Protophloem Formation
Molecular genetic framework for protophloem formation Antia Rodriguez-Villalon, Bojan Gujas, Yeon Hee Kang, Alice S. Breda, Pietro Cattaneo, Stephen Depuydt, and Christian S. Hardtke1 Department of Plant Molecular Biology, University of Lausanne, CH-1015 Lausanne, Switzerland Edited by Philip N. Benfey, Duke University, Durham, NC, and approved June 20, 2014 (received for review April 22, 2014) The phloem performs essential systemic functions in tracheo- eventually differentiates into metaphloem further up in the root phytes, yet little is known about its molecular genetic specifica- to functionally replace the protophloem (1), which differentiates tion. Here we show that application of the peptide ligand closer to the tip from the cell file emerging from the outer cell. CLAVATA3/EMBRYO SURROUNDING REGION 45 (CLE45) specifi- Finally, as these cell files differentiate into sieve elements, the cally inhibits specification of protophloem in Arabidopsis roots by immediately neighboring cell files (which originate from differ- locking the sieve element precursor cell in its preceding develop- ent stem cells) synchronously differentiate into companion cells, mental state. CLE45 treatment, as well as viable transgenic expres- although with a delay of two to four cells with respect to the sieve sion of a weak CLE45G6T variant, interferes not only with com- elements. In the Arabidopsis root meristem, the protophloem mitment to sieve element fate but also with the formative sieve strands can be identified in confocal microscopy through their element precursor cell division that creates protophloem and enhanced propidium iodide cell wall staining (Fig. 1B) (4). metaphloem cell files. However, the absence of this division Mutant analyses in Arabidopsis have identified various genes in- appears to be a secondary effect of discontinuous sieve element volved in the formation of vascular tissues (1, 3, 5–9). -

Lecture 6 (I) Phloem Tissue Or Bast Or Leptome
Lecture 6 (i) Phloem tissue or bast or Leptome • Phloem is another type of conducting tissue like xylem which is responsible for conduction of organic substances. • The phloem from the procambium is called primary phloem and that formed from vascular cambium is called secondary phloem. • The phloem is composed of four elements: Sieve tube elements, Companion cells, Phloem parenchyma (Bast parenchyma) and Phloem fibers (Bast fibers). (i) Sieve tube elements Sieve tubes are tube-like structures, composed of elongated cells, arranged in longitudinal series and associated with companion cells. Their walls are thin and made of cellulose. In a mature sieve tube the nucleus is absent but cytoplasm as well as large vacuole is present. Although without nucleus, it is living and the nucleus of the companion cells controls its functional activities. The transverse partition walls are perforated by a number of pores, giving the appearance of sieves. They are called the sieve plates. A sieve plate is called simple if it has only one sieve area. A sieve plate is called compound if it has many sieve areas. At the end of the growing season the sieve plate is covered by a deposit of carbohydrate called callose. But in the spring, when the active season begins, it gets dissolved; in old sieve tubes callose forms a permanent deposit. The P proteins (P-phloem) are seen evenly distributed throughout the lumen of the sieve tube. During wounding, along with callose, P-proteins help in sealing. In lower vascular plants and gymnosperms in place of sieve tube elements, sieve cells are present. -

Stelar System and Its Evolution
Dr Rajib Kagyung Stellar theory and its evolution STELAR SYSTEM AND ITS EVOLUTION 10.1 WHAT ARE STELE AND STELLAR THEORY? The central cylinder or core of vascular tissue, consisting of xylem, phloem, pericycle and sometimes medullary rays and pith, is technically called stele. Van Tieghem and Douliot (1886) developed “Stellar Theory”. They used the term stele in collective sense and mentioned that the stele is not only made up of xylem and phloem, but the tissue like pericycle, vascular rays and pith are also associated with it. According to them the cortex and the stele are the fundamental parts of a shoot and both these parts are separated from each other by the endodermis. According to stellar theory, primarily there is no fundamental difference in the gross anatomy of stem and roots, because in both of them a stele is surrounded by the cortex is present. Although stele is real entity and present universally in all axes of the plants, I n higher vascular plants like ferns, gymnosperms and angiosperms the leaf traces are large and it plays an important role in the vascular system of the axis. Foster and Gifford (1959) have mentioned that the most debated and controversial aspect of “stellar theory is the nature of the anatomical boundaries which separate the cortex from the stele”. According to Van Tieghem and Douliot (1886) the endodermis represents the inner boundary of the cortex. The cells of the endodermal layer have the characteristic casparian strip strips. But in the stems of many seed plants, the characteristic endodermal layer is not present.